Angiostrongylus cantonensis an Atypical Presenilin: Epitope Mapping, Characterization, and Development of an ELISA Peptide Assay for Specific Diagnostic of Angiostrongyliasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Rat Infected Sera and Ethics Statement
2.2. Synthesis of the Cellulose-Membrane-Bound Peptide Array
2.3. Screening of SPOT Membranes
2.4. Scanning and Measurement of Spot Signal Intensities
2.5. Synthesis of Peptides and Preparation of the MAPs
2.6. ELISA-Peptide
2.7. Database Searches, Computational, and Phylogeny Studies
2.8. Ethics Statement
2.9. Statistical Analysis
3. Results
3.1. Identification and Mapping of Linear Epitopes Using Synthetic Peptides
3.2. Absence of Crucial Motifs for A22 Aspartyl Protease Family
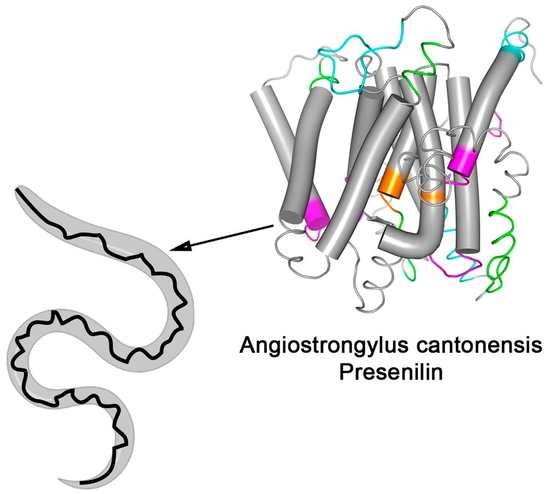
3.3. Spatial Location of the Most Reactive Epitopes
3.4. Phylogeny and Protein-Protein Interaction
3.5. Selection of Putative Specific Epitopes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiao-Ping, W.; De-Hua, L.; Xing-Quan, Z.; Xiao-Guang, C.; Zhao-Rong, L. Human angiostrongyliasis. Lancet Infect. Dis. 2008, 8, 621–630. [Google Scholar] [CrossRef]
- Tesana, S.; Srisawangwong, T.; Sithithaworn, P.; Laha, T.; Andrews, R. Prevalence and intensity of infection with third-stage larvae of Angiostrongylus cantonensis in mollusks from Northeast Thailand. Am. J. Trop. Med. Hyg. 2009, 80, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Maldonado-Junior, A.; Mora, J.; Morassutti, A.; Rodriguez, R.; Solano-Barquero, A.; Tijerino, A.; Vargas, M.; Graef-Teixeira, C. Abdominal angiostrongyliasis in the Americas: Fifty years since the discovery of a new metastrongylid species, Angiostrongylus costaricensis. Parasites Vectors 2021, 14, 374. [Google Scholar] [CrossRef] [PubMed]
- Federspiel, F.; Skovmand, S.; Skarphedinsson, S. Eosinophilic meningitis due to Angiostrongylus cantonensis in Europe. Int. J. Infect. Dis. 2020, 93, 28–39. [Google Scholar] [CrossRef]
- Center for Disease Control and Prevention. Available online: https://www.cdc.gov/parasites/angiostrongylus/epi.html (accessed on 11 December 2021).
- Robles, G.; Loría, R.; Lobo, F.; Robles, A.; Valle, S.; Cordero, C. Granuloma eosinofílico parasitario intestinal. Rev. Méd. Hosp. Nac. Niñ. 1968, 3, 67–80. [Google Scholar]
- Morera, P. Life history and redescription of Angiostrongylus costaricensis Moreira and Céspedes. Am. J. Trop. Med. Hyg. 1973, 22, 613–621. [Google Scholar] [CrossRef]
- Céspedes, R.; Salas, J.; Mekbel, S.; Troper, L.; Mullner, F.; Morera, P. Granulomas entéricos y linfáticos con intensa eosinoflia tisular producidos por un estrongilídeo (Strongylata; Railliet y Henry, 1913) I. Patol. Acta Med. Costarr. 1967, 10, 235–255. [Google Scholar]
- Chen, H.T. A new pulmonary nematode of rats, Pulmonema cantonensis ng, nsp from Canton. Ann. Parasitol. 1935, 13, 312–317. [Google Scholar] [CrossRef] [Green Version]
- Barratt, J.; Chan, D.; Sandaradura, I.; Malik, R.; Spielman, D.; Lee, R.; Marriott, D.; Harkness, J.; Ellis, J.; Stark, D. Angiostrongylus cantonensis: A review of its distribution, molecular biology, and clinical significance as a human pathogen. Parasitology 2016, 143, 1087–1118. [Google Scholar] [CrossRef] [Green Version]
- Jarvi, S.I.; Jacob, J.; Sugihara, R.T.; Leinbach, I.L.; Klasner, I.H.; Kaluna, L.M.; Snook, K.A.; Howe, M.K.; Jacquier, S.H.; Lange, I.; et al. Validation of a death assay for Angiostrongylus cantonensis larvae (L3) using propidium iodide in a rat model (Rattus norvegicus). Parasitology 2019, 146, 1421–1428. [Google Scholar] [CrossRef] [Green Version]
- Tsai, H.C.; Lee, S.S.J.; Huang, C.K.; Yen, C.M.; Chen, E.R.; Liu, Y.C. Outbreak of eosinophilic meningitis associated with drinking raw vegetable juice in southern Taiwan. Am. J. Trop. Med. Hyg. 2004, 71, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Tang, P.; Yen, C.M.; Chow, K.P.; Wang, L.C. A transcriptomic analysis on gene expressions in the infective third and pathogenic fifth larval stages of Angiostrongylus cantonensis. Parasitol. Int. 2014, 63, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Walker, M.D.; Zunt, J.R. Neuroparasitic infections: Cestodes, Trematodes, and Protozoans. Semin. Neurol. 2005, 25, 262–277. [Google Scholar] [CrossRef] [Green Version]
- Graeff-Teixeira, C.; Morassutti, A.L.; Jones, M.K. Diagnosing and understanding angiostrongyliasis, a zoonotic cause of meningitis. ACS Chem. Neurosci. 2018, 9, 393–394. [Google Scholar] [CrossRef] [Green Version]
- Eamsobhana, P.; Yong, H.S. Immunological diagnosis of human angiostrongyliasis due to Angiostrongylus cantonensis (Nematoda: Angiostrongylidae). Int. J. Infect. Dis. 2009, 13, 425–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eamsobhana, P. Eosinophilic meningitis caused by Angiostrongylus cantonensis: A neglected disease with escalating importance. Trop. Biomed. 2014, 31, 569–578. [Google Scholar]
- Wang, Q.P.; Wu, Z.D.; Wei, J.; Owen, R.L.; Lun, Z.R. Human Angiostrongylus cantonensis: An update. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, P.P.; Qvarnstrom, Y.; Whelen, A.C.; Saucier, C.; da Silva, A.J.; Eamsobhana, P. The current status of laboratory diagnosis of Angiostrongylus cantonensis infections in humans using serologic and molecular methods. Hawaii J. Med. Public Health 2013, 72, 55–57. [Google Scholar] [PubMed]
- Chen, S.N. Enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies to Angiostrongylus cantonensis. Trans. R. Soc. Trop. Med. Hyg. 1986, 80, 398–405. [Google Scholar] [CrossRef]
- Cross, J.H.; Chi, J.C. ELISA for the detection of Angiostrongylus cantonensis antibodies in patients with eosinophilic meningitis. Southeast Asian J. Trop. Med. Public Health 1982, 13, 73–76. [Google Scholar]
- Zoua, Y.; Guanb, H.; Wu, H.; Bu, H.; Yan, L.; Zhu, Y.; He, J. Angiostrongyliasis detected by next-generation sequencing in ELISA-negative eosinophilic meningitis: A case report. Int. J. Infect. Dis. 2020, 97, 177–179. [Google Scholar] [CrossRef] [PubMed]
- Eamsobhana, P.; Ongrotchanakun, J.; Yoolek, A.; Punthuprapasa, P.; Monkong, N.; Dekumyoy, P. Multi-immunodot for rapid differential diagnosis of eosinophilic meningitis due to parasitic infections. J. Helminthol. 2006, 80, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Eamsobhana, P.; Gan, X.; Ma, A.; Wang, Y.; Wanachiwanawin, D.; Yong, H. Dot immunogold filtration assay (DIGFA) for the rapid detection of specific antibodies against the rat lungwormAngiostrongylus cantonensis(Nematoda: Metastrongyloidea) using purified 31-kDa antigen. J. Helminthol. 2014, 88, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Maleewong, W.; Sombatsawat, P.; Intapan, P.M.; Wongkham, C.; Chotmongkol, V. Immunoblot evaluation of the specificity of the 29-kDa antigen from young adult female worms Angiostrongylus cantonensis for immunodiagnosis of human angiostrongyliasis. Asian Pac. J. Allergy Immunol. 2001, 19, 267–273. [Google Scholar]
- Somboonpatarakun, C.; Intapan, P.M.; Sanpool, O.; Wongkham, C.; Maleewong, W. Application of recombinant Angiostrongylus cantonensis galectin-2 protein for serodiagnosis of human Angiostrongyliasis by immunoblotting. Am. J. Trop. Med. Hyg. 2019, 101, 851–858. [Google Scholar] [CrossRef] [PubMed]
- Intapan, P.M.; Maleewong, W.; Sawanyawisuth, K.; Chotmongkol, V. Evaluation of human IgG subclass antibodies in the serodiagnosis of angiostrongyliasis. Parasitol. Res. 2003, 89, 425–429. [Google Scholar] [CrossRef]
- Morassutti, A.L.; Levert, K.; Perelygin, A.; da Silva, A.J.; Wilkins, P.; Graeff-Teixeira, C. The 31-kDa antigen of Angiostrongylus cantonensis comprises distinct antigenic glycoproteins. Vector Borne Zoonotic Dis. 2012, 12, 961–968. [Google Scholar] [CrossRef] [Green Version]
- Eamsobhana, P.; Tungtrongchitr, A.; Wanachiwanawin, D.; Yong, H.S. Immunochromatographic test for rapid serological diagnosis of human angiostrongyliasis. Int. J. Infect. Dis. 2018, 73, 69–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somboonpatarakun, C.; Intapan, P.M.; Sadaow, L.; Rodpai, R.; Sanpool, O.; Maleewong, W. Development of an immunochromatographic device to detect antibodies for rapid diagnosis of human angiostrongyliasis. Parasitology 2020, 147, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Graeff-Teixeira, C.; Pascoal, V.F.; Rodriguez, R.; Morassutti, A.L.; Intapan, P.M.; Maleewong, W. Abdominal angiostrongyliasis can be diagnosed with an immunochromatographic rapid test with recombinant galactin from Angiostrongylus cantonensis. Mem. Inst. Oswaldo Cruz 2020, 115, e200201. [Google Scholar] [CrossRef] [PubMed]
- Morassutti, A.; Rascoe, L.; Handali, S.; Da Silva, A.; Wilkins, P.; Graeff-Teixeira, C. Cross-reactivity of the 31 kDa antigen of Angiostrongylus cantonensis–Dealing with the immunodiagnosis of meningoencephalitis. Parasitology 2017, 144, 459–463. [Google Scholar] [CrossRef]
- Cognato, B.B.; Handali, S.; de Mattos Pereira, L.; Barradas, J.R.; Januário da Silva, A.; Graeff-Teixeira, C.; Morassutti, A.L. Identification of cross-reactive markers to strengthen the development of immunodiagnostic methods for angiostrongyliasis and other parasitic infections. Exp. Parasitol. 2020, 218, 107999. [Google Scholar] [CrossRef]
- Xie, M.; Zhou, Z.; Guo, S.; Li, Z.; Zhao, H.; Deng, J. Next-generation sequencing specifies Angiostrongylus eosinophilic meningoencephalitis in infants: Two case reports. Medicine 2019, 98, e16985. [Google Scholar] [CrossRef]
- Feng, L.; Zhang, A.; Que, J.; Zhou, H.; Wang, H.; Guan, Y.; Shen, C.; Sun, X.; Lai, R.; Peng, F.; et al. The metagenomic next-generation sequencing in diagnosing central nervous system angiostrongyliasis: A case report. BMC Infect. Dis. 2020, 20, 691. [Google Scholar] [CrossRef] [PubMed]
- Morassutti, A.L.; Levert, K.; Pinto, P.M.; da Silva, A.J.; Wilkins, P.; Graeff-Teixeira, C. Characterization of Angiostrongylus cantonensis excretory-secretory proteins as potential diagnostic targets. Exp. Parasitol. 2012, 130, 26–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdel-Rahman, H.M.; Kimura, T.; Hidaka, K.; Kiso, A.; Nezami, A.; Freire, E.; Hayashi, Y.; Kiso, Y. Design of inhibitors against HIV, HTLV-I, and Plasmodium falciparum aspartic proteases. Biol. Chem. 2004, 385, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, S.; Dalton, J.P.; Loukas, A. Proteases in helminth- and allergen-induced inflammatory responses. Chem. Immunol. Allergy 2006, 90, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Zawrotniak, M.; Bochenska, O.; Karkowska-Kuleta, J.; Seweryn-Ozog, K.; Aoki, W.; Ueda, M.; Kozik, A.; Rapala-Kozik, M. Aspartic proteases and major cell wall components in Candida albicans trigger the release of neutrophil extracellular traps. Front. Cell. Infect. Microbiol. 2017, 7, 414. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulou, A.A.; Fluhrer, R. Signaling functions of intramembrane aspartyl-proteases. Front. Cardiovasc. Med. 2020, 7, 591787. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.; Ribeiro Carlton, S.M.; Simões, I. Atypical, and nucellin-like aspartic proteases: Emerging players in plant developmental processes and stress responses. J. Exp. Bot. 2019, 70, 2059–2076. [Google Scholar] [CrossRef]
- Soares, A.; Niedermaier, S.; Faro, R.; Loos, A.; Manadas, B.; Faro, C.; Huesgen, P.F.; Cheung, A.Y.; Simões, I. An atypical aspartic protease modulates lateral root development in Arabidopsis thaliana. J. Exp. Bot. 2019, 70, 2157–2171. [Google Scholar] [CrossRef]
- Guo, Y.; Tu, T.; Zheng, J.; Ren, Y.; Wang, Y.; Bai, Y.; Su, X.; Wang, Y.; Yao, B.; Huang, H.; et al. A novel thermostable aspartic protease from Talaromyces leycettanus and its specific autocatalytic activation through an intermediate transition state. Appl. Microbiol. Biotechnol. 2020, 104, 4915–4926. [Google Scholar] [CrossRef]
- Silva, F.R.; Napoleão-Pêgo, P.; De-Simone, S.G. Identification of linear B epitopes of pertactin of Bordetella pertussis induced by immunization with whole and acellular vaccine. Vaccine 2014, 32, 6251–6258. [Google Scholar] [CrossRef] [Green Version]
- De-Simone, S.G.; Souza, A.L.A.; Melgarejo, A.R.; Aguiar, A.S.; Provance, D.W., Jr. Development of elisa assay to detect specific human IgE anti-therapeutic horse sera. Toxicon 2017, 138, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Prado, I.C.; Provance, D.W., Jr.; De-Simone, S.G. Ultrasensitive and rapid immuno-detection of human IgE hypersensitive anti-therapeutic horse sera using an electrochemical immunosensor. Anal. Biochem. 2017, 538, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Bottino, C.G.; Gomes, L.P.; Zauza, P.L.; Pereira, J.B.; Coura, J.R.; De-Simone, S.G. Chagas disease-specific antigens: Characterization of CRA/FRA epitopes by synthetic peptide mapping and evaluation on ELISA-peptide assay. BMC Infect. Dis. 2013, 13, 568. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, K.; Stoffel, W. TMbase-A database of membrane-spanning protein segments. Biol. Chem. Hoppe Seyler 1993, 347, 166. [Google Scholar]
- Bernse, A.; Viklund, H.; Hennerdal, A.; Elofsson, A. TOPCONS: Consensus prediction of membrane protein topology. Nucleic Acids Res. 2009, 37, W465–W468. [Google Scholar] [CrossRef] [Green Version]
- Hönigschmid, P.; Frishman, D. Accurate prediction of helix interactions and residue contacts in membrane proteins. J. Struct. Biol. 2016, 194, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bohm, C.; Dodd, R.; Chen, F.; Qamar, S.; Schmitt-Ulms, G.; Fraser, P.E.; St George-Hyslop, P.H. Structural biology of presenilin 1 complexes. Mol. Neurodegener. 2014, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lechuga, G.C.; Bottino, C.C.G.; Pinho, R.T.; Souza, A.L.A.; Provance, D.W., Jr.; De-Simone, S.G. Trypanosoma cruzi presenilin-like transmembrane aspartyl protease: Characterization and cellular localization. Biomolecules 2020, 10, 1564. [Google Scholar] [CrossRef]
- Lechuga, G.C.; Napoleão-Pêgo, P.; Gomes, L.R.; Durans, A.M.; Provance, D.W., Jr.; De-Simone, S.G. Nicastrin-like, a novel transmembrane protein from Trypanosoma cruzi associated to the flagellar pocket. Microorganisms 2021, 9, 1750. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Yao, L.L.; Song, Z.M.; Li, X.P.; Hua, Q.Q.; Li, Q.; Pan, C.W.; Xia, C.M. Development-specific differences in the proteomics of Angiostrongylus cantonensis. PLoS ONE 2013, 8, e76982. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.Y.; Lu, P.J.; Cheng, C.J.; Jhan, K.Y.; Yeh, S.C.; Wang, L.C. Proteomic analysis of excretory-secretory products from young adults of Angiostrongylus cantonensis. Mem. Inst. Oswaldo Cruz. 2019, 114, e180556. [Google Scholar] [CrossRef] [PubMed]
- Yücel, S.S.; Lemberg, M.K. Signal peptide peptidase-type proteases: Versatile regulators with functions ranging from limited proteolysis to protein degradation. J. Mol. Biol. 2020, 432, 5063–5078. [Google Scholar] [CrossRef]
- Mentrup, T.; Cabrera-Cabrera, F.; Fluhrer, R.; Schröder, B. Physiological functions of SPP/SPPL intramembrane proteases. Cell. Mol. Life Sci. 2020, 77, 2959–2979. [Google Scholar] [CrossRef] [Green Version]
- Dehury, B.; Tang, N.; Kepp, K.P. Insights into membrane-bound presenilin 2 from all-atom molecular dynamics simulations. J. Biomol. Struct. Dyn. 2020, 38, 3196–3210. [Google Scholar] [CrossRef]
- Struhl, G.; Greenwald, I. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 229–234. [Google Scholar] [CrossRef]
- Hwang, K.P.; Chang, S.H.; Wang, L.C. Alterations in the expression level of a putative aspartic protease in the development of Angiostrongylus cantonensis. Acta Trop. 2010, 113, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Hutton, M.; Nyborg, A.; Baker, M.; Jansen, K.; Golde, T.E. Identification of a novel family of presenilin homologues. Hum. Mol. Genet. 2002, 11, 1037–1044. [Google Scholar] [CrossRef] [Green Version]
- Raut, P.; Glass, J.B.; Lieberman, R.L. Archaeal roots of intramembrane aspartyl protease siblings signal peptide peptidase and presenilin. Proteins 2021, 89, 232–241. [Google Scholar] [CrossRef]
- Tomita, T.; Watabiki, T.; Takikawa, R.; Morohashi, Y.; Takasugi, N.; Kopan, R.; De Strooper, B.; Iwatsubo, T. The first Proline of PALP motif at the C terminus of presenilins is obligatory for stabilization, complex formation, and gamma-secretase activities of presenilins. J. Biol. Chem. 2001, 276, 33273–33281. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Beher, D.; Nyborg, A.C.; Shearman, M.S.; Golde, T.E.; Goate, A. C-terminal PAL motif of PS and PS homologs required for normal active site conformation. J. Neurochem. 2006, 96, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Honarnejad, K.; Herms, J. Presenilins: Role in calcium homeostasis. Int. J. Biochem. Cell Biol. 2012, 44, 1983–1986. [Google Scholar] [CrossRef]
- Mehra, R.; Kepp, K.P. Identification of structural calcium-binding sites in membrane-bound presenilin 1 and 2. J. Phys. Chem. B 2020, 124, 4697–4711. [Google Scholar] [CrossRef] [PubMed]
- Dries, D.R.; Yu, G. Assembly, maturation and trafficking of the gamma-secretase complex in Alzheimer’s disease. Curr. Alzheimer Res. 2008, 5, 132–146. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Sato, K.; Liou, W.; Pant, S.; Harada, A.; Grant, B.D. Regulation of endocytic recycling by C. elegans Rab35 and its regulator RME-4, a coated-pit protein. EMBO J. 2008, 27, 1183–1196. [Google Scholar] [CrossRef] [Green Version]
- Sarasija, S.; Norman, K.R. A γ-Secretase independent role for presenilin in calcium homeostasis impacts mitochondrial function and morphology in Caenorhabditis elegans. Genetics 2015, 201, 1453–1466. [Google Scholar] [CrossRef] [Green Version]
- Oikawa, N.; Walter, J. Presenilins and γ-secretase in membrane proteostasis. Cells 2019, 8, 209. [Google Scholar] [CrossRef] [Green Version]






| Code | Epitope | Position aa Number | Secondary Structure 1 | Cross-Reactivity Sequence 2 (UniProtKB) | Soluble Synthesized Peptide Sequences 3 |
|---|---|---|---|---|---|
| PsAg1 | TISTTARNSKAASTN | 36–50 | C | NQP | TISTTARNSKAASTN |
| PsAg2 | VKRLYGPTDM | 56–65 | C | NQP | GSGVKRLYGPTDMGG |
| PsAg3 | GVNLIPTPYVIYNEP | 91–105 | H + C + H | NQP | GVNLIPTPYVIYNEP |
| PsAg4 | TKLLHA | 111–116 | H | NQP | TKLLHAVANAATFLV |
| PsAg5 | QQIL | 167–170 | H | NQP | GSGFSFVQYQQILGG |
| PsAg6 | SIFWKGPMRLQQASL | 196–210 | C + H | NQP | SIFWKGPMRLQQASL |
| PsAg7 | PQWTS | 216–230 | C + H | NQP | TVTLTIMQILPQWTS |
| PsAg8 | PSSGRFIESFRMPSL | 306–320 | C | NQP | PSSGRFIESFRMPSL |
| PsAg9 | ERNIRLGLG | 326–334 | C | NQP | ERNIRLGLGDFIFYS |
| PsAg10 | VYLNRSWFHS | 359–366 | H | NQP | GSGVYLNRSWFHSGG |
| PsAg11 | TSKGTASTTNIRHIW | 374–388 | H + C + H | NQP | TSKGTASTTNIRHIW |
| PsAg12 | KFAVELNR | 401–408 | C | NQP | SSRFAMSKFAVELNR |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De-Simone, S.G.; Napoleão-Pêgo, P.; Gonçalves, P.S.; Lechuga, G.C.; Mandonado, A., Jr.; Graeff-Teixeira, C.; Provance, D.W., Jr. Angiostrongylus cantonensis an Atypical Presenilin: Epitope Mapping, Characterization, and Development of an ELISA Peptide Assay for Specific Diagnostic of Angiostrongyliasis. Membranes 2022, 12, 108. https://doi.org/10.3390/membranes12020108
De-Simone SG, Napoleão-Pêgo P, Gonçalves PS, Lechuga GC, Mandonado A Jr., Graeff-Teixeira C, Provance DW Jr. Angiostrongylus cantonensis an Atypical Presenilin: Epitope Mapping, Characterization, and Development of an ELISA Peptide Assay for Specific Diagnostic of Angiostrongyliasis. Membranes. 2022; 12(2):108. https://doi.org/10.3390/membranes12020108
Chicago/Turabian StyleDe-Simone, Salvatore G., Paloma Napoleão-Pêgo, Priscila S. Gonçalves, Guilherme C. Lechuga, Arnaldo Mandonado, Jr., Carlos Graeff-Teixeira, and David W. Provance, Jr. 2022. "Angiostrongylus cantonensis an Atypical Presenilin: Epitope Mapping, Characterization, and Development of an ELISA Peptide Assay for Specific Diagnostic of Angiostrongyliasis" Membranes 12, no. 2: 108. https://doi.org/10.3390/membranes12020108