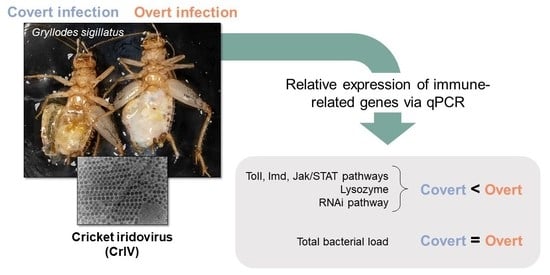
Induction of Multiple Immune Signaling Pathways in Gryllodes sigillatus Crickets during Overt Viral Infections
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cricket Colonies
2.2. Viral Imaging Via Electron Microscopy
2.3. RNA Extraction and cDNA Synthesis
2.4. Gene Target-Specific Primer Design
2.5. Reverse Transcriptase Quantitative PCR (RT-qPCR) Detection and Quantification
2.6. Statistical Analysis
3. Results
3.1. TEM Imaging
3.2. Immune Signaling Pathways
3.3. RNAi Pathway
3.4. Lysozyme
3.5. Microbial Load
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Defoliart, G.R. Edible Insects as Minilivestock. Biodivers. Conserv. 1995, 4, 306–321. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Addison, M.F.; Terblanche, J.S. Mass-rearing of insects for pest management: Challenges, synergies and advances from evolutionary physiology. Crop. Prot. 2012, 38, 87–94. [Google Scholar] [CrossRef]
- Bosch, G.; Zhang, S.; Oonincx, D.G.A.B.; Hendriks, W.H. Protein quality of insects as potential ingredients for dog and cat foods. J. Nutr. Sci. 2014, 3, E29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makkar, H.P.S.; Tran, G.; Heuzé, V.; Ankers, P. State-of-the-art on use of insects as animal feed. Anim. Feed. Sci. Technol. 2014, 197, 1–33. [Google Scholar] [CrossRef]
- van Huis, A.; Van Itterbeeck, J.; Klunder, H.; Mertens, E.; Halloran, A.; Muir, G.; Vantomme, P. Edible Insects: Future Prospects for Food and Feed Security; FAO Forestry Paper 171; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2013; Volume 171. [Google Scholar]
- Hahn, T.; Tafi, E.; Paul, A.; Salvia, R.; Falabella, P.; Zibek, S. Current state of chitin purification and chitosan production from insects. J. Chem. Technol. Biotechnol. 2020, 95, 2775–2795. [Google Scholar] [CrossRef]
- Surendra, K.C.; Tomberlin, J.K.; van Huis, A.; Cammack, J.A.; Heckmann, L.H.L.; Khanal, S.K. Rethinking organic wastes bioconversion: Evaluating the potential of the black soldier fly (Hermetia illucens (L.))(Diptera: Stratiomyidae)(BSF). J. Waste Manag. 2020, 117, 58–80. [Google Scholar] [CrossRef]
- Ortiz, J.C.; Ruiz, A.T.; Morales-Ramos, J.A.; Thomas, M.; Rojas, M.G.; Tomberlin, J.K.; Yi, L.; Han, R.; Giroud, L.; Jullien, R.L. Chapter 6—Insect Mass Production Technologies, in Insects as Sustainable Food Ingredients. In Insects as Sustainable Food Ingredients; Dossey, A.T., Morales-Ramos, J.A., Rojas, M.G., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 153–201. ISBN 978-0-12-802856-8. [Google Scholar]
- Bertola, M.; Mutinelli, A.F. Systematic Review on Viruses in Mass-Reared Edible Insect Species. Viruses 2021, 13, 2280. [Google Scholar] [CrossRef]
- Maciel-Vergara, G.; Jensen, A.B.; Lecocq, A.; Eilenberg, J. Diseases in edible insect rearing systems. J. Insects Food Feed. 2021, 7, 621–638. [Google Scholar] [CrossRef]
- Maciel-Vergara, G.; Ros, V.I. Viruses of insects reared for food and feed. J. Invertebr. Pathol. 2017, 147, 60–75. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Granberg, F.; Onorati, P.; Jansson, A.; Berggren, Å. Virus prospecting in crickets—Discovery and strain divergence of a novel iflavirus in wild and cultivated Acheta domesticus. Viruses 2021, 13, 364. [Google Scholar] [CrossRef]
- Duffield, K.R.; Hunt, J.; Sadd, B.M.; Sakaluk, S.K.; Oppert, B.; Rosario, K.; Behle, R.W.; Ramirez, J.L. Active and Covert Infections of Cricket Iridovirus and Acheta domesticus Densovirus in Reared Gryllodes sigillatus Crickets. Front. Microbiol. 2021, 12, 780796. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, J.R.; Granberg, F.; Low, M.; Onorati, P.; Semberg, E.; Jansson, A.; Berggren, Å. Virus Diversity and Loads in Crickets Reared for Feed: Implications for Husbandry. Front. Vet. Sci. 2021, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Weissman, D.B.; Gray, D.A.; Pham, H.T.; Tijssen, P. Billions and billions sold: Pet-feeder crickets (Orthoptera: Gryllidae), commercial cricket farms, an epizootic densovirus, and government regulations make for a potential disaster. Zootaxa 2012, 3504, 67–88. [Google Scholar] [CrossRef]
- Sorrell, I.; White, A.; Pedersen, A.B.; Hails, R.S.; Boots, M. The evolution of covert, silent infection as a parasite strategy. Proc. R. Soc. B 2009, 276, 2217–2226. [Google Scholar] [CrossRef]
- Adamo, S.A.; Parsons, N.M. The emergency life-history stage and immunity in the cricket, Gryllus texensis. Anim. Behav. 2006, 72, 235–244. [Google Scholar] [CrossRef]
- Reginald, K.; Wong, Y.R.; Shah, S.M.R.; Teh, K.F.; Jalin, E.J.F.; Khan, N.A. Investigating immune responses of the house cricket, Acheta domesticus to pathogenic Escherichia coli K1. Microbes Infect 2021, 23, 104876. [Google Scholar] [CrossRef]
- Cho, Y.; Cho, S. Hemocyte-hemocyte adhesion by granulocytes is associated with cellular immunity in the cricket, Gryllus bimaculatus. Sci. Rep. 2019, 9, 18066. [Google Scholar] [CrossRef] [Green Version]
- Drayton, J.M.; Jennions, M.D. Inbreeding and measures of immune function in the cricket Teleogryllus commodus. Behav. Ecol. 2011, 22, 486–492. [Google Scholar] [CrossRef] [Green Version]
- Adamo, S.A.; Jensen, M.; Younger, M. Changes in lifetime immunocompetence in male and female Gryllus texensis (formerly G. integer): Trade-offs between immunity and reproduction. Anim. Behav. 2001, 62, 417–425. [Google Scholar] [CrossRef]
- Hampton, K.J.; Duffield, K.R.; Hunt, J.; Sakaluk, S.K.; Sadd, B.M. Male and female genotype and a genotype-by-genotype interaction mediate the effects of mating on cellular but not humoral immunity in female decorated crickets. Heredity 2021, 126, 477–490. [Google Scholar] [CrossRef]
- Piñera, A.V.; Charles, H.M.; Dinh, T.A.; Killian, K.A. Maturation of the immune system of the male house cricket, Acheta domesticus. J. Insect Physiol. 2013, 59, 752–760. [Google Scholar] [CrossRef]
- Ferguson, L.V.; Dhakal, P.; Lebenzon, J.E.; Heinrichs, D.E.; Bucking, C.; Sinclair, B.J. Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct. Ecol. 2018, 32, 2357–2368. [Google Scholar] [CrossRef] [Green Version]
- Duffield, K.R.; Hampton, K.J.; Houslay, T.M.; Hunt, J.; Sadd, B.M.; Sakaluk, S.K. Inbreeding alters context-dependent reproductive effort and immunity in male crickets. J. Evol. Biol. 2019, 32, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Duffield, K.R.; Hampton, K.J.; Houslay, T.M.; Rapkin, J.; Hunt, J.; Sadd, B.M.; Sakaluk, S.K. Macronutrient intake and simulated infection threat independently affect life history traits of male decorated crickets. Ecol. Evol. 2020, 10, 11766–11778. [Google Scholar] [CrossRef] [PubMed]
- Kerr, A.M.; Gershman, S.N.; Sakaluk, S.K. Experimentally induced spermatophore production and immune responses reveal a trade-off in crickets. Behav. Ecol. 2010, 21, 647–654. [Google Scholar] [CrossRef]
- Letendre, C.; Duffield, K.R.; Sadd, B.M.; Sakaluk, S.K.; House, C.M.; Hunt, J. Genetic covariance in immune measures and pathogen resistance in decorated crickets is sex and pathogen specific. J. Anim. Ecol. 2022, 91, 1471–1488. [Google Scholar] [CrossRef]
- Gershman, S.N.; Barnett, C.A.; Pettinger, A.M.; Weddle, C.B.; Hunt, J.; Sakaluk, S.K. Inbred decorated crickets exhibit higher measures of macroparasitic immunity than outbred individuals. Heredity 2010, 105, 282–289. [Google Scholar] [CrossRef] [Green Version]
- Simmons, L.W. Resource allocation trade-off between sperm quality and immunity in the field cricket, Teleogryllus oceanicus. Behav. Ecol. 2011, 23, 168–173. [Google Scholar] [CrossRef] [Green Version]
- Adamo, S.A. Estimating disease resistance in insects: Phenoloxidase and lysozyme-like activity and disease resistance in the cricket Gryllus texensis. J. Insect Physiol. 2004, 50, 209–216. [Google Scholar] [CrossRef]
- Bascuñán-García, A.P.; Lara, C.; Córdoba-Aguilar, A. Immune investment impairs growth, female reproduction and survival in the house cricket, Acheta domesticus. J. Insect Physiol. 2010, 56, 204–211. [Google Scholar] [CrossRef]
- Park, Y.; Stanley, D. Physiological trade-off between cellular immunity and flight capability in the wing-dimorphic sand cricket, Gryllus firmus. J. Asia Pac. Entomol. 2015, 18, 553–559. [Google Scholar] [CrossRef]
- Duffield, K.R.; Hampton, K.J.; Houslay, T.M.; Hunt, J.; Rapkin, J.; Sakaluk, S.K.; Sadd, B.M. Age-dependent variation in the terminal investment threshold in male crickets. Evolution 2018, 72, 578–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, A.; Lee, T.Y.M.; McMillan, L.E.; Easy, R.; Adamo, S.A. Immunity for nothing and the eggs for free: Apparent lack of both physiological trade-offs and terminal reproductive investment in female crickets (Gryllus texensis). PLoS ONE 2019, 14, e0209957. [Google Scholar] [CrossRef] [PubMed]
- Sorrell, M.R.; Killian, K.A. Innate immune system function following systemic RNA-interference of the Fragile X Mental Retardation 1 gene in the cricket Acheta domesticus. J. Insect Physiol. 2020, 126, 104097. [Google Scholar] [CrossRef]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [Green Version]
- Casanova-Torres, Á.M.; Goodrich-Blair, H. Immune signaling and antimicrobial peptide expression in Lepidoptera. Insects 2013, 4, 320–338. [Google Scholar] [CrossRef] [Green Version]
- Sim, S.; Jupatanakul, N.; Dimopoulos, G. Mosquito immunity against arboviruses. Viruses 2014, 6, 4479–4504. [Google Scholar] [CrossRef] [Green Version]
- Gottar, M.; Gobert, V.; Michel, T.; Belvin, M.; Duyk, G.; Hoffmann, J.A.; Ferrandon, D.; Royet, J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002, 416, 640–644. [Google Scholar] [CrossRef]
- Horng, T.; Medzhitov, R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 12654–12658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.; Khanuja, B.S.; Ip, Y.T. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes Dev. 1999, 13, 792–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choe, K.M.; Werner, T.; Stoven, S.; Hultmark, D.; Anderson, K.V. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 2002, 296, 359–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silverman, N.; Zhou, R.; Stöven, S.; Pandey, N.; Hultmark, D.; Maniatis, T. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000, 14, 2461–2471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Small, S.; Desplan, C.; Dearolf, C.R.; Darnell Jr, J.E. Identification of a Stat gene that functions in Drosophila development. Cell 1996, 84, 421–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betz, A.; Lampen, N.; Martinek, S.; Young, M.W.; Darnell Jr, J.E. A Drosophila PIAS homologue negatively regulates stat92E. Proc. Natl. Acad. Sci. USA 2001, 98, 9563–9568. [Google Scholar] [CrossRef] [Green Version]
- Feng, M.; Fei, S.; Xia, J.; Labropoulou, V.; Swevers, L.; Sun, J. Antimicrobial peptides as potential antiviral factors in insect antiviral immune response. Front. Immunol. 2020, 11, 2030. [Google Scholar] [CrossRef]
- Schneider, P.M. Purification and properties of three lysozymes from hemolymph of the cricket, Gryllus bimaculatus (De Geer). Insect Biochem. 1985, 15, 463–470. [Google Scholar] [CrossRef]
- Chen, T.T.; Tan, L.R.; Hu, N.; Dong, Z.Q.; Hu, Z.G.; Jiang, Y.M.; Chen, P.; Pan, M.H.; Lu, C. C-lysozyme contributes to antiviral immunity in Bombyx mori against nucleopolyhedrovirus infection. J. Insect Physiol. 2018, 108, 54–60. [Google Scholar] [CrossRef]
- Wang, X.H.; Aliyari, R.; Li, W.X.; Li, H.W.; Kim, K.; Carthew, R.; Atkinson, P.; Ding, S.W. RNA interference directs innate immunity against viruses in adult Drosophila. Science 2006, 312, 452–454. [Google Scholar] [CrossRef]
- Paradkar, P.N.; Trinidad, L.; Voysey, R.; Duchemin, J.B.; Walker, P.J. Secreted Vago restricts West Nile virus infection in Culex mosquito cells by activating the Jak-STAT pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 18915–18920. [Google Scholar] [CrossRef] [Green Version]
- Rand, T.A.; Ginalski, K.; Grishin, N.V.; Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. USA 2004, 101, 14385–14389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T. Covert iridovirus infection of blackfly larvae. Proc. R. Soc. Lond. B Biol. Sci. 1993, 251, 225–230. [Google Scholar]
- Williams, T. Natural invertebrate hosts of iridoviruses (Iridoviridae). Neotrop. Entomol. 2008, 37, 615–632. [Google Scholar] [CrossRef] [Green Version]
- Tonka, T.; Weiser, J. Iridovirus infection in mayfly larvae. J. Invertebr. Pathol. 2000, 76, 229–231. [Google Scholar] [CrossRef] [PubMed]
- Jupatanakul, N.; Sim, S.; Dimopoulos, G. The insect microbiome modulates vector competence for arboviruses. Viruses 2014, 6, 4294–4313. [Google Scholar] [CrossRef] [PubMed]
- Bozzola, J.; Russell, L. Electron Microscopy; Jones and Bartlett Publishers: Boston, MA, USA, 1992; ISBN 9780867201260. [Google Scholar]
- Foquet, B.; Rapkin, J.; Sharma, M.D.; Sadd, B.M.; Sakaluk, S.K.; Hunt, J. Transcriptomic responses of females to consumption of nuptial food gifts as a potential mediator of sexual conflict in decorated crickets. J. Evol. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform 2009, 10, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.L.; Barrett, T.; Benson, D.A.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Edgar, R.; Federhen, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2007, 36, D13–D21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Ugene Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xi, Z.; Ramirez, J.L.; Dimopoulos, G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS Pathog. 2008, 4, e1000098. [Google Scholar] [CrossRef]
- Costa, A.; Jan, E.; Sarnow, P.; Schneider, D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 2009, 4, e7436. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.L.; Souza-Neto, J.; Torres Cosme, R.; Rovira, J.; Ortiz, A.; Pascale, J.M.; Dimopoulos, G. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 2012, 6, e1561. [Google Scholar] [CrossRef] [PubMed]
- Oppert, B.; Perkin, L.C.; Lorenzen, M.; Dossey, A.T. Transcriptome analysis of life stages of the house cricket, Acheta domesticus, to improve insect crop production. Sci. Rep. 2020, 10, 3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.H.; Markkandan, K.; Kim, I.W.; Kim, S.Y.; Seo, M.; Kim, M.A.; Kim, S.H.; Hwang, J.S. De novo assembly and functional annotation of the emma field cricket (Teleogryllus emma) transcriptome. J. Asia Pac. Entomol. 2019, 22, 1–5. [Google Scholar] [CrossRef]
- Johnston, P.R.; Paris, V.; Rolff, J. Immune gene regulation in the gut during metamorphosis in a holo- versus a hemimetabolous insect. Philos. Trans. R Soc. B 2019, 374, 20190073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Chen, J.; Keyhani, N.O.; Zhang, Z.; Li, S.; Xia, Y. Comparative transcriptomic analysis of immune responses of the migratory locust, Locusta migratoria, to challenge by the fungal insect pathogen, Metarhizium acridum. BMC Genom. 2015, 16, 867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamo, S.A.; Kovalko, I.; Easy, R.H.; Stoltz, D. A viral aphrodisiac in the cricket Gryllus texensis. J. Exp. Biol. 2014, 217, 1970–1976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, X.-J.; Ling, E.; Yu, X.-Q. The role of lysozyme in the prophenoloxidase activation system of Manduca sexta: An in vitro approach. Dev. Comp. Immunol. 2010, 34, 264–271. [Google Scholar] [CrossRef] [Green Version]
- Papp, T.; Marschang, R.E. Detection and characterization of invertebrate iridoviruses found in reptiles and prey insects in Europe over the past two decades. Viruses 2019, 11, 600. [Google Scholar] [CrossRef] [Green Version]
- Bronkhorst, A.W.; van Cleef, K.W.; Venselaar, H.; van Rij, R.P. A dsRNA-binding protein of a complex invertebrate DNA virus suppresses the Drosophila RNAi response. Nucleic Acids Res. 2014, 42, 12237–12248. [Google Scholar] [CrossRef] [Green Version]
- Bronkhorst, A.W.; van Cleef, K.W.; Vodovar, N.; İnce, İ.A.; Blanc, H.; Vlak, J.M.; Saleh, M.C.; van Rij, R.P. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA 2012, 109, E3604–E3613. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA interference–mediated antiviral immunity and virus-specific inducible responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- West, C.; Rus, F.; Chen, Y.; Kleino, A.; Gangloff, M.; Gammon, D.B.; Silverman, N. IIV-6 Inhibits NF-κB Responses in Drosophila. Viruses 2019, 11, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.; Virto, C.; Murillo, R.; Caballero, P. Covert infection of insects by baculoviruses. Front. Microbiol. 2017, 8, 1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, T.; Ylla, G.; Extavour, C.G. Genomics and genome editing techniques of crickets, an emerging model insect for biology and food science. Curr. Opin. Insect Sci. 2022, 50, 100881. [Google Scholar] [CrossRef]




| Target | Gene ID | Primer Sequence | Amplicon Size (bp) | Efficiency | R2 |
|---|---|---|---|---|---|
| Dorsal | ON081012 | GGTAGGGGCTCTCTTTGGTC | 107 | 98% | 0.9978 |
| CGTTCTGCTGGCTCTATTCC | |||||
| Dif | ON081011 | TATGAATGCGAAGGGAGGTC | 130 | 98% | 0.9963 |
| ACAGCACGACCCTGATAACC | |||||
| Cactus | ON081013 | GTGTGACCAGCGTAAGTGGA | 75 | 92% | 0.9979 |
| CCTCAGCAGTGTGTTGCATT | |||||
| MyD88 | ON081014 | AACGGCTCCAGCATCTAAAA | 115 | 90% | 0.9949 |
| TGGTGGATCTGTCAAGCAAG | |||||
| PGRP-LC | ON081023 | AATAGCCAGAGGAGCAGCAA | 99 | 100% | 0.9982 |
| GGCCAAACTGGAGATACCAA | |||||
| Imd | ON081024 | ATTCCTCGCATCAACACTCC | 143 | 96% | 0.9839 |
| TCAGGTGATGGTGATTTGGA | |||||
| Relish | ON081022 | GGCAGTTTCACCTTCCACAT | 118 | 96% | 0.9999 |
| GCTGCAGATGGCTCTAAAGG | |||||
| STAT5B | ON081015 | GCCCCATACCATGTCCTAGA | 109 | 91% | 0.9971 |
| TATGTGCACAATCCCCTCAA | |||||
| PIAS | ON081016 | GGTCACAAAGCCTTCAGGAG | 82 | 100% | 0.9973 |
| AGTTCTCTGGACGTGCCAAT | |||||
| Domeless | ON081017 | CCATTCAGGCACCAGAAGAT | 124 | 99% | 0.9995 |
| TGCCAAAAGAACCAGTTTCC | |||||
| Argonaute-2 | ON081018 | TGCATGTTCATCCCTTGAAA | 135 | 95% | 0.9976 |
| GTTCCCGGCAAGACATTAAA | |||||
| Dicer-2 | ON081020 | CCCTTTCTCCATGACTTCCA | 78 | 100% | 0.9992 |
| CCTCCAATTTTCAGCACCAC | |||||
| R2D2 | ON081019 | ATGTCTGCCTGTTGGGAAAC | 99 | 99% | 0.9986 |
| GCGCTCACGTGTACTGTTGT | |||||
| Lysozyme | ON081021 | TTACGACTACGGCCTGTTCC | 84 | 98% | 0.9994 |
| TCGCACTTCATCTTGCAATC | |||||
| 18S rRNA | KR904053 | GCCGTTCTTAGTTCGTGGAG | 130 | 97% | 0.9979 |
| CGCCTGTCCCTCTAAGAAGA | |||||
| 16S rRNA | AF514593 | TCGTCACCCCAACCAAATAC | 106 | 96% | 0.9984 |
| TAATGGGGGACGAGAAGACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duffield, K.R.; Foquet, B.; Stasko, J.A.; Hunt, J.; Sadd, B.M.; Sakaluk, S.K.; Ramirez, J.L. Induction of Multiple Immune Signaling Pathways in Gryllodes sigillatus Crickets during Overt Viral Infections. Viruses 2022, 14, 2712. https://doi.org/10.3390/v14122712
Duffield KR, Foquet B, Stasko JA, Hunt J, Sadd BM, Sakaluk SK, Ramirez JL. Induction of Multiple Immune Signaling Pathways in Gryllodes sigillatus Crickets during Overt Viral Infections. Viruses. 2022; 14(12):2712. https://doi.org/10.3390/v14122712
Chicago/Turabian StyleDuffield, Kristin R., Bert Foquet, Judith A. Stasko, John Hunt, Ben M. Sadd, Scott K. Sakaluk, and José L. Ramirez. 2022. "Induction of Multiple Immune Signaling Pathways in Gryllodes sigillatus Crickets during Overt Viral Infections" Viruses 14, no. 12: 2712. https://doi.org/10.3390/v14122712