Photophore Distribution and Enzymatic Diversity Within the Photogenic Integument of the Cookie-Cutter Shark Isistius brasiliensis (Chondrichthyes: Dalatiidae)
- 1Biology of Marine Organisms and Biomimetics, Research Institute for Biosciences, University of Mons (UMONS), Mons, Belgium
- 2Marine Biology Laboratory, Earth and Life Institute, University of Louvain (UCLouvain), Louvain-la-Neuve, Belgium
The cookie-cutter shark Isistius brasiliensis (Squaliformes: Dalatiidae) is a deep-sea species that emits a blue luminescence ventrally, except at the level of a black band located beneath the jaw. This study aims to (i) investigate the distribution and histology of the photophores (i.e., light-emitting organs) along the shark body, (ii) describe the tissue-specific transcriptomes of the black band integument region (i.e., non-photogenic) and the ventral integument region (i.e., photogenic), (iii) describe the repertoire of enzyme-coding transcripts expressed the two integument regions, and (iv) analyze the potential expression of transcripts coding for luciferase-like enzymes (i.e., close homologs of known luciferases involved in the bioluminescence of other organisms). Our analyses confirm the black band’s non-photogenic status and photophore absence within this region. The sub-rostral area is the region where the photophore density is the highest. In parallel, paired-end Illumina sequencing has been used to generate two pilot transcriptomes, from the black band and the ventral integument tissues of one individual. In total, 68,943 predicted unigenes have been obtained (i.e., 64,606 for the black band transcriptome, 43,996 for the ventral integument transcriptome) with 43,473 unigenes showing significant similarities to known sequences from public databases. BLAST search analyses of known luciferases, coupled with comparative predicted gene expression (i.e., photogenic versus non-photogenic), support the hypothesis that the species uses an unknown luciferase system. An enzymatic repertoire was predicted based on the PRIAM database, and Enzyme Commission numbers were assigned for all detected enzyme-coding unigenes. These pilot transcriptomes based on a single specimen, and the predicted enzyme repertoire, constitute a valuable resource for future investigations on the biology of this enigmatic luminous shark.
Introduction
Chondrichthyes (i.e., sharks, skates, rays, and chimeras) are among the most abundant and diverse vertebrate taxa (Carrier et al., 2012; Ebert et al., 2013). Among all described shark species (i.e., 537), 47% are totally or partially living in the deep-sea environment (i.e., below 200 m) (Kyne and Simpfendorfer, 2007; Cotton and Grubbs, 2015; Pollerspöck and Straube, 2019). Deep-water sharks are mainly represented in eight orders: Echinorhiniformes (100% of deep-sea species), Squaliformes (86%), Hexanchiformes (86%), Pristiorhiformes (50%), Carcharhiniformes (40%), Lamniformes (20%), Squatiniformes (8%), and Orectolobiformes (4%) (Pollerspöck and Straube, 2019).
At the level of the skin epidermis, and mainly ventrally, some deep-sea Squaliforme sharks possess well developed light organs (i.e., photophores) able to intrinsically emit a blue-green light (Claes and Mallefet, 2009; Renwart et al., 2014, 2015; Claes et al., 2015; Duchatelet et al., 2019a). Among Squaliformes, three families (out of the seven) comprise bioluminescent species (i.e., able to emit light): the Etmopteridae, the Dalatiidae and the Somniosidae (Claes and Mallefet, 2009; Straube et al., 2015; Duchatelet et al., 2021). Shark luminescence is assumed to be mainly used for counterillumination purposes in Dalatiidae and Somniosidae, while Etmopteridae have developed an intricate pattern of luminescent areas suggested to be used for diverse functions such as counterillumination, aposematism, and intraspecific communication (Claes and Mallefet, 2008; Claes et al., 2010, 2013, 2015; Duchatelet et al., 2019b, 2021; Mallefet et al., 2021). 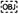 Although Somniosidae (i.e., Zameus squamulosus) luminescence remains largely enigmatic, the light emission as well as the presence and histology of photophores have been described recently (Straube et al., 2015; Duchatelet et al., 2021).
Although Somniosidae (i.e., Zameus squamulosus) luminescence remains largely enigmatic, the light emission as well as the presence and histology of photophores have been described recently (Straube et al., 2015; Duchatelet et al., 2021).
In addition to its not so common “bioluminescence function,” shark skin is also known to be a pleiotropic tissue involved in a variety of functions such as senses (e.g., Fields, 2007; Hart and Collin, 2014), protection and hydrodynamics through the placoid scale squamation pattern (e.g., Wainwright et al., 1978; Reif, 1985; Meyer and Seegers, 2012; Oeffner and Lauder, 2012), immunity (e.g., Moore et al., 1993; Tsutsui et al., 2015), and color changes for camouflage or UV protection through melanophore pigment motion (e.g., Lowe and Goodman-Lowe, 1996; Visconti et al., 1999; Robbins and Fox, 2012).
The cookie-cutter shark, Isistius brasiliensis (Squaliformes, Dalatiidae) is one rare luminous shark found in the bathy- and mesopelagic zone of the temperate and tropical waters (Jahn and Haedrich, 1987; Ebert et al., 2013; Figure 1A). With a maximum length of about 50 cm, I. brasiliensis migrates from the ocean’s deep layer to the surface at night, feeding mainly on squids and small fishes (Papastamatiou et al., 2010). This shark also presents an occasional ectoparasite lifestyle, feeding on a round-shaped chunk of flesh it removes from various larger pelagic organisms such as bony fishes (e.g., Jones, 1971; Muñoz-Chápuli et al., 1988; Papastamatiou et al., 2010), sharks (e.g., Hoyos-Padilla et al., 2013), whales (e.g., Dwyer and Visser, 2011; Feunteun et al., 2018; Murakami et al., 2018) and seals (Le Boeuf et al., 1987). The cookie-cutter shark emits blue light (Claes et al., 2015). On its photogenic ventral side, between the gill slits and the pectoral fins, the cookie-cutter shark has a well-marked black band (also referred to as dark collar) that does not produce luminescence (Widder, 1998). Although encounters with this elusive animal are increasing, little is known about its bioluminescence and the associated biological functions. Information lacks concerning potential similitudes (e.g., photophore morphology, light emission control) with other investigated bioluminescent sharks (e.g., Squaliolus aliae another Dalatiidae species studied by Claes et al., 2011 and Duchatelet et al., 2020a). As for all luminous sharks, the luminous compounds (i.e., luciferin and luciferase or photoprotein) underlying the light production remain unknown in this organism.
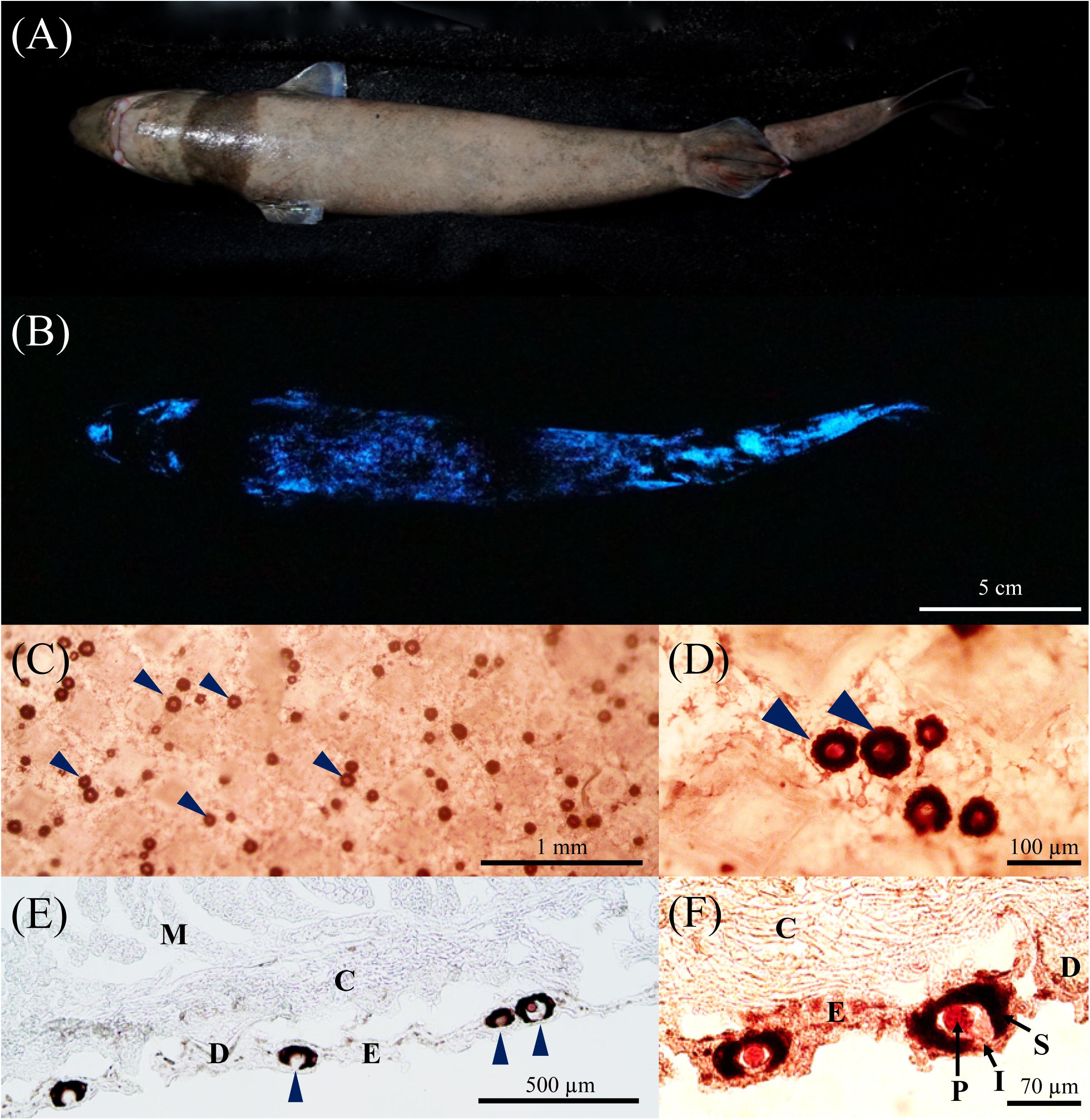
Figure 1. Bioluminescence of Isistius brasiliensis. (A) Ventral side of I. brasiliensis. The dark-collar or black band is visible on the anterior part. (B) The ventral luminous pattern observed on the ventral side. An absence of bioluminescence is observed on the dark collar. The dark region in the middle of the shark is artifactual and corresponds to the area where the specimen was stabilized for the picture. (C,D) In vivo observation of the photophores present on the ventral skin region. (E,F) Observation of the photophores using classical histology. Arrows indicate the epidermal photophores. Legend, C, connective tissue; D, dermal denticle; E, epidermis; M, muscles; I, iris-like structure; P, photocyte; S, pigmented sheath.
In the present study, the morphology of light-emitting photophores and their distribution along the body were investigated. In parallel, the analyses of transcriptomes of the ventral integument, the photogenic region, and the non-photogenic black band integument region allowed us to highlight the diversity of enzyme-coding transcripts expressed within these functionally different tissues, with a special emphasis on the search for luciferase/photoprotein candidates.
Materials and Methods
Specimen Sampling
Adult male specimens of Isistius brasiliensis (n = 3) were captured as bycatch by longliner fisherman in La Réunion Island (n = 2; registration number IS_Bmar_001 and 002) and by trawling at 1,000 m depth during the “Sampling the Abyss” cruise1 in June 2017 (n = 1; registration number NMV A 31829-001, Victoria Museums) operating along the coast of Australia and Tasmania. Living cookie-cutter shark from Australia was maintained onboard in small tank filled with cold seawater before being euthanized by deep sedation in clove oil. The specimen collected in Australia (NMV A 31829-001, Australia) was used for in vivo observations. Integument tissues of the ventral luminous area and the black band of the same specimen were used for a pilot transcriptomic approach. In parallel, skin tissues of two individuals (IS_Bmar_001 and 002), collected in La Réunion, were used for the histological study. Due to the scarcity of this shark and the technical difficulties to isolate the epidermis from the rest of the integument layers, samples contained the upper photophore-bearing part of the skin (i.e., epidermis) as well as underlying tissues (i.e., dermis, subcutis and underlying muscles). These transcriptomes will be referred to as ventral and black band integument transcriptomes.
High Sensitivity Macrophotography and Histology
Freshly collected individuals of I. brasiliensis were used for luminous areas detection using high sensitivity macrophotography performed in the dark. In parallel, skin patches of different areas along the body (i.e., rostral, mandibular, black band, pectoral, ventral, ventrolateral, dorsolateral, dorsal, dorsal fin, pelvic, caudal areas) were observed under a light microscope and photographed. Photophore density per area was evaluated by counting the number of photophores per square millimeter (n = 40 fields per area). Photophore diameter was also measured for each investigated area.
Patches of ventral and black band integument (1 cm2) were dissected and fixed in 4% paraformaldehyde in phosphate buffer saline (PBS) for 1 day at 4°C, then stored at 4°C in PBS for light microscopy analyses. Skin patches were bathed in PBS with increasing concentrations of sucrose (10% for 1 h, 20% for 1 h, and 30% overnight), before being embedded in O. C. T. compound (Optimal Cutting Temperature compound, Sakura, Zoeterwude, Netherlands) and quickly frozen at −80°C. Sections, 10 μm in thickness, were obtained using a CM3050 S. Leica cryostat microtome (Germany), mounted on clean glass slides, and left overnight to dry. Slides were observed with a microscope (Leitz Diaplan, Germany) equipped with a ToupCam camera (UCMOS Series C-mount USB2.0 CMOS camera, ToupTek, Zhejiang, China).
RNA Extraction, cDNA Library Preparation, and Sequencing and RNA-Seq Analyses
Tissues of ventral and black band integument from one individual were dissected and frozen in liquid nitrogen. Shark tissues were then permeabilized in RNAlaterTM-Ice (Life Technologies) overnight at −20°C, then stored at −80°C until RNA extraction procedure. Extraction of total RNA content was performed following the Trizol reagent-based protocol. RNA extract qualities were assessed by gel electrophoresis on a 1.2% TAE agarose gel, as well as by spectrophotometry using a Nanodrop spectrometer (LabTech International). RNA quality was also checked by size-exclusion chromatography with an Agilent Technologies 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). The rarity of the species imposed this limited sampling (n = 1).
cDNA library preparation and sequencing were performed by the Beijing Genomics Institute (BGI, Hong Kong) according to the Illumina protocol (Illumina, San Diego, CA, United States). Illumina HiSeq X Ten platform was used for high-throughput sequencing to generate 150-bp paired-end reads. Raw sequences were cleaned following different filtering steps: removal of reads (i) containing the adaptor sequence only; (ii) containing over 5% of unknown nucleotides; (iii) comprising more than 20% of bases with a quality value lower than 10. Read cleaning was performed using filter_fq (BGI internal software) with default settings. The remaining filtered reads were used to generate a reference de novo transcriptome assembly for I. brasiliensis, derived from the black band and ventral integument tissues. The FastQC program (Andrews, 2010) was used to assess the quality of the reads.
Assembly of the de novo transcriptome of I. brasiliensis was performed via short paired-end reads using the Trinity software (Grabherr et al., 2011; version release-20121005; min_contig_length 100, group_pairs_distance 250, path_reinforcement_distance 95, min_kmer_cov 2). Distinct sequences (unigenes) were obtained, after Trinity assembly, using the TGI Clustering Tool (TGICL) (Pertea et al., 2003) following a procedure described in Delroisse et al. (2015, 2016, 2018). The unigenes here form either a group in which the similarity between overlapping sequences is greater than 94%, or singleton which correspond to single unigenes (Das et al., 2016). As the length of sequences assembled is a recognized criterion for assembly success in terms of contiguity, we calculated the size distribution of both contigs and unigenes.
Based on the global unigene assembly, all the clean reads of each sample were mapped to the unigene dataset using the Bowtie2 software (v2.2.5) (Langmead and Salzberg, 2012). The gene expression levels were calculated using RSEM (v1.2.12) (Li and Dewey, 2011). Unigene expression was expressed through the “Fragments per kilobase of transcript, per million fragments sequenced” (FPKM) values as described in Delroisse et al. (2015, 2016). It must be clarified that the transcriptome data have been generated in the purpose of new gene discovery, not differential expression analyses, as no biological or technical replication was performed in the framework of this study. For descriptive purposes, we identified “differentially expressed genes (DEGs)” between the two transcriptomes using the “PoissonDis” method that is based on the Poisson distribution (Audic and Claverie, 1997). Unigenes for which the absolute value of log2 (Foldchange) was higher or equal to 1 and with a corrected P-value FDR inferior or equal to 0.001 were considered as DEGs.
Functional Gene Annotation and Transcriptome Completeness Analyses
All generated unigenes were used for homology searches against protein databases (NCBI NR, Swissprot, KOG, KEGG, Interpro) using BLASTx analyses (v2.2.23, E-value < 1e–5). Best results were selected to annotate the unigenes. When the results from different databases were conflicting, the results from the NR database (i.e., the most complete) were preferentially selected, followed by Swissprot, KEGG and COG databases. Unigene sequences were also compared to nucleotide databases NT (non-redundant NCBI nucleotide database, E-value < 1e–5, BLASTn). The best aligning results of the BLASTx alignments between unigenes and protein databases like NCBI NR, Swiss-Prot, KEGG, and COG were used to identify unigenes’ sequence direction. When results from different databases are conflicting, the priority order NR, Swiss-Prot, KEGG, and COG was followed to decide on sequence direction for unigenes. The Blast2GO program (v2.5.0) was used with NR annotation to get GO annotation according to molecular function, biological process, and cellular component ontologies2. To further annotate the unigenes, the web Platform FunctionAnnotator3 was specifically used to annotate enzymes-coding predicted transcripts based on the PRIAM databases (E-value < 1e–10).
Transcriptome completeness was assessed using the bioinformatics tool BUSCO v4 (Benchmarking Universal Single-Copy Orthologs) to obtain the percentage of single-copy orthologs represented in the dataset Vertebrata_odb9. BUSCO analyses and result visualization were performed in the Galaxy platform4.
Luciferase-Like Candidate Searches
To identify potential luciferase/photoprotein candidates in the transcriptome of the luminous shark I. brasiliensis, reference luciferase/photoprotein sequences from various luminous organisms (e.g., Porifera, Cnidaria, Ctenophora, Insecta, Crustacea, Echinodermata), obtained in open-access NCBI online databases were used in a “tBLASTn/reciprocal BLASTx.” Reference sequences, corresponding to biochemically tested enzymes or putative enzymes described in the literature, are listed in the Supplementary Table 1. It has to be stated that some reference luciferase candidates have yet to be confirmed biochemically (e.g., Arachnocampa luminosa luciferase, Suberites domuncula luciferase, Amphiura filiformis luciferase, Watasenia scintillans luciferase; Supplementary Table 1) although strong indications suggested their involvement in the light emission of the considered organisms (Müller et al., 2009; Delroisse et al., 2014, 2017a,b; Gimenez et al., 2016; Watkins et al., 2018). Homologous sequences to reference luciferases/photoproteins were first searched using tBLASTn analyses in the newly generated integument transcriptome of I. brasiliensis. Candidate matches were then used as queries in a reciprocal BLASTx search against the NCBI NR online database to highlight sequences with high similarity with potential luciferases.
Results
High Sensitivity Photography and Histological Analyses
High sensitivity macrophotographs recorded from the cookie-cutter shark in the dark revealed a diffuse blue glow present all over the ventral body side except at the level of the caudal fin and the area between the jaws and the pectoral fins (i.e., the so-called black band). No other specific pattern (e.g., sexual pattern at the pelvic area, flank marking, dorsal lines) was observed. Luminescence was absent from the dorsal side (dorsal area or dorsal fins) in the tested specimen (n = 1) (Figure 1B).
In vivo observations of the different skin regions zones allowed us to identify and visualize photophores (Figures 1C,D) and to measure their density according to the other skin areas (Supplementary Figure 1). Photophores (mean diameter = 56.1 ± 5.4 μm; no statistical difference between areas), that are scattered between the pavement-like placoid scales (Figure 1C), are present in large amount in all the studied areas except for the black band, dorsal, supra-rostral, supra-mandibular and dorsal fin areas, where only few light organs per square millimeters have been observed (0.68 ± 0.47; 6.28 ± 0.14, 6.58 ± 0.74, 3.64 ± 0.33, 1.23 ± 0.41 photophore mm–2, respectively, Supplementary Figure 1). Comparatively, ventral, sub-mandibular and sub-rostral areas present a photophore density of 28.75 ± 0.51, 28.40 ± 1.47 and 45.83 ± 2.17 photophore mm–2, respectively (Supplementary Figure 1). A photophore density gradient (mainly dorso-ventral, but also antero-posterior) is clearly observed along the shark body (Supplementary Figure 1).
Transverse sections through the photogenic ventral skin of I. brasiliensis revealed the internal structure of the photophores (Figures 1E,F). Similarly to the photophores described by Seigel (1978) for the genus Squaliolus, I. brasiliensis photophores are located in the stratified squamous epidermis and are composed of a single photogenic cell (i.e., photocyte) embedded in a cup-shaped pigmented sheath, surmounted by a few cells forming an iris-like structure (Figure 1F). Lens cells, topping the light organ, are observed in some sections.
De novo Transcriptome Sequencing, Transcriptome Quality Assessment and Annotation
A total of 88.17 and 99.60 million raw reads, with a length of 150 bp, were generated from the black band and ventral integument libraries, respectively. The datasets of raw reads were deposited in NCBI database: Bioproject PRJNA648842 (under SRA experiment numbers SRR12351835 and SRR12351836). After low quality reads filtering, 68.96 and 80.35 million high quality reads (10.34 and 12.05 Gbases in total), obtained were used to assemble the black band and ventral integument transcriptomes with the Trinity software. Clean read quality across all bases is presented in Supplementary Figure 2. Q20 percentages (i.e., base quality more than 20) for the clean reads were higher to 78.2% for both transcriptome datasets. Contigs were generated following the overlapping information of high-quality reads. The mean contig lengths were of 660 and 605 bp and the N50 (i.e., the length of the longest contig such that all contigs of at least that length represent at least 50% of the bases of the assembly) were 1,431 and 1,154 bp for the black band and ventral integument transcriptomic data, respectively.
Contigs were further assembled into unigenes, i.e., unique non-redundant sequences, using paired end joining and gap filling. A total of 64,606 unigenes were obtained for the black band dataset and 43,996 for the ventral integument dataset, for a total of 68,943 different unigenes. For the unigenes, the N50 were 2,023 and 1,661 bp for the black band and ventral integument transcriptomes, respectively. The unigenes length distribution is presented in the Figure 2A. A N50 of 2,115 is reached for the global unigene dataset. The global unigene dataset includes 14,786 clusters and 54,157 singletons.
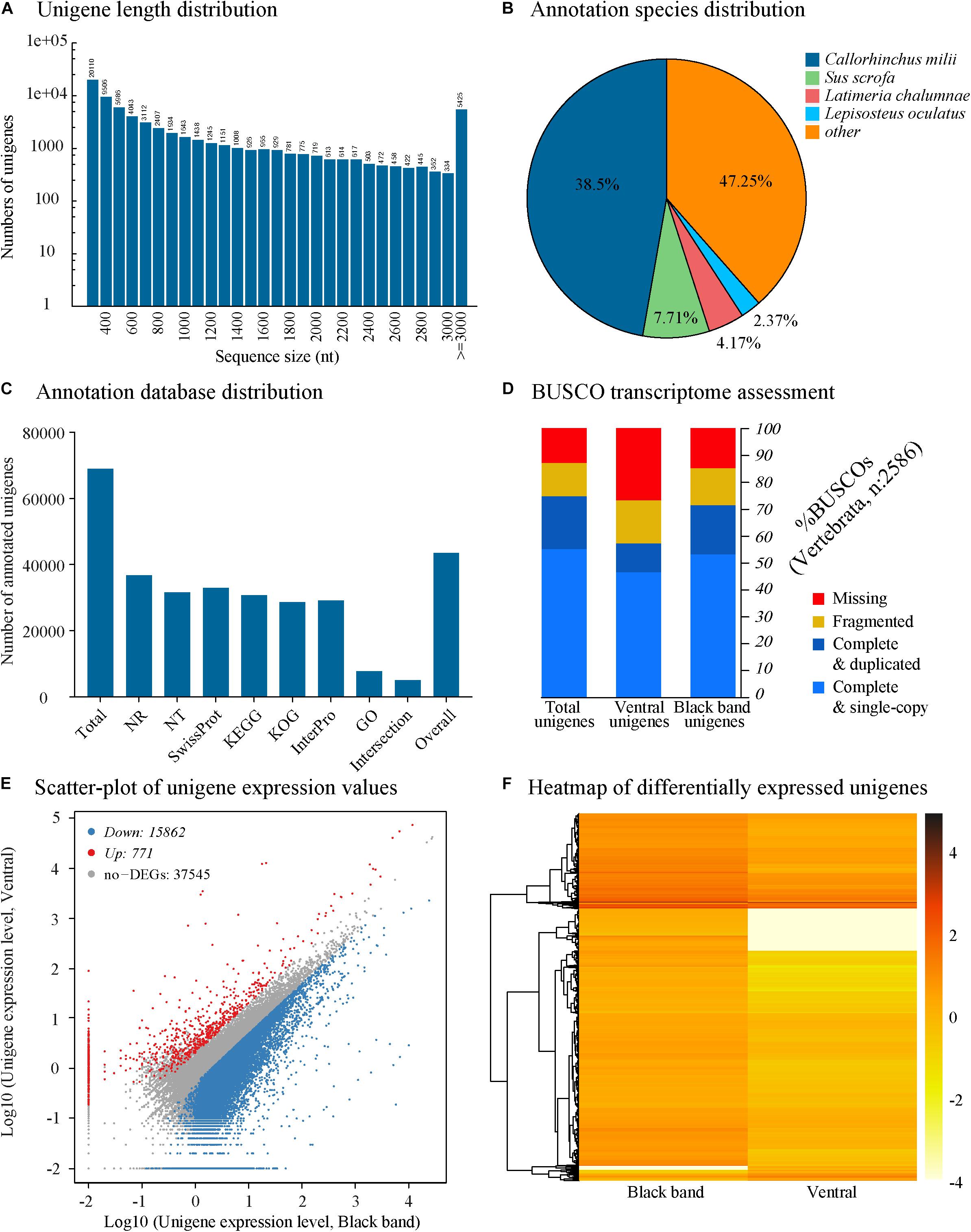
Figure 2. Isistius brasiliensis transcriptome summary. (A) Unigenes length distribution of the global transcriptome, (B) species distribution of the top BLAST hits for all unigenes, (C) annotation summary for all investigated databases: NR, KOG, KEGG, Swissprot and Interpro. The category “intersection” refers to unigenes that are commonly annotated in all databases. In contrast, the category “overall” refers to unigenes that are uniquely annotated in one of the tested databases (D) BUSCO transcriptome assessment, (E) comparative unigenes expression between the black band and the ventral integument transcriptomes, (F) heatmap of log2 (FoldChange) of differentially expressed unigenes based on the Poisson distribution (corrected P-value FDR ≤ 0.001, | log2(FoldChange)| ≥ 1) (Audic and Claverie, 1997).
On all the 68,943 I. brasiliensis pooled unigenes, 43,474 (63.06%) show significant matches to the NCBI NR database (Supplementary Table 2, Sheet A “Unigene annotation”). Because of the lack of genome reference in I. brasiliensis and, possibly, the relatively short length of some unigenes, around 37% of the assembled sequences could not be matched to any known genes. Among annotated unigenes from the pooled transcriptome, 38.5% of the sequences were matched to the elephant shark Callorhinchus milii for which the genome has been sequenced (Figure 2B). On the 68,943 I. brasiliensis unigenes present in the reference transcriptome, 43,473 show significant matches to public databases (E-value threshold > 1e–5): 36,785 to NR (53.36%), 31,628 to NT (45.88%), 32,965 to SwissProt (47.81%), 30,727 to KEGG (41.49%), 28,602 to KOG (41.49%), 29,168 to InterPro (42.31%) and 7,747 to GO (11.24%) (Figure 2C).
To evaluate the transcriptome’s completeness, a BUSCO search of 2,583 vertebrate gene groups resulted in 74.6% of complete orthologs (including 55% of single-copy BUSCOs and 19.6% of duplicated BUSCOs) for the global unigenes dataset. Separate analyses of both transcriptomes (i.e., ventral versus black band integuments) revealed a lower level of completeness of the ventral integument transcriptome (Figure 2D).
All the clean reads from each sample were mapped to the global unigene dataset, and the gene expression levels were calculated. On a total of 54,178 mapped unigenes, 16,147 were only detected in the black band integument transcriptome and 3,544, only, in the ventral integument transcriptome while 34,487 were detected in both transcriptomes. For descriptive purposes, a comparative gene expression analysis was performed by plotting the unigenes expression values (i.e., log10[unigenes normalized count for the black band integument transcriptome] against log10[unigenes normalized count for the ventral integument transcriptome]), calculated for all predicted unigenes (Figure 2E). However, as stated in the methodology, no biological or technical replication was performed as a part of the study and the present datasets have been generated in the purpose of new gene discovery only. Based on the “| log2FoldChange| ≥ 1” threshold and the corrected P-value FDR ≤ 0.001, 15,862 unigenes were found to be “upregulated” in the black band integument transcriptome and 771 in the ventral integument transcriptome (Figures 2E,F).
Within the 20 most expressed unigenes of the ventral and black band integument transcriptomes (Supplementary Table 2 Sheets B, C: “MostExpressed Ventral” and “MostExpressed BlackB”), predicted genes such as “Glyceraldehyde-3-phosphate dehydrogenase,” “Fructose-bisphosphate aldolase A” are specifically represented. Both predicted transcripts coding for enzymes involved in the glycolytic pathway are possibly expressed within the muscular layer present below the skin layers. Several genes coding for muscle protein actors are well represented such as “Actin alpha cardiac muscle 1-like,” “Myosin regulatory light chain 2, skeletal muscle isoform,” “Tropomyosin alpha-1 chain isoform X1,” “creatine kinase M-type,” “Sarcoplasmic/endoplasmic reticulum calcium ATPase1” and various myosins. Other potential muscle markers are highly expressed in the ventral integument transcriptome (e.g., “Beta-enolase”) or in the black band transcriptome (e.g., “Telethonin”). Globally, it indicates a strong representation of muscle-specific genes expressed within the underlying muscle layer, possibly due to a facilitated mRNA extraction from muscle tissue compared to other tissue types from the integument.
Comparative unigene expression analyses, based on the FPKM value comparisons, indicates that within the 20 most differentially expressed unigenes in the ventral integument transcriptome (Supplementary Table 2, Sheet D: “MostDiffExpressed Ventral”), many muscle specific markers can also be found (“myosin-4 isoform X1,” “myosin regulatory light chain 2, skeletal muscle isoform,” “myosin-binding protein C, fast-type isoform X1,” “myosin-binding protein H-like isoform X2,” “tropomodulin 4, muscle,” “troponin T, cardiac muscle isoforms-like isoform X1,” “myosin, heavy chain 2, skeletal muscle, adult,” “CAVP-target proteins”). Other unigenes correspond to “2-acylglycerol O-acyltransferase 2-like,” “immunoglobulin-like and fibronectin type III domain-containing protein 1 isoform X2,” “parvalbumin, thymic,” “immunoglobulin-like and fibronectin type III domain-containing protein 1 isoform X1” and “PDZ and LIM domain protein 5 isoform X11.” Conversely, other muscle-specific markers are found within the 20 most differentially expressed unigenes in the black band transcriptome such as “tropomyosin alpha-1 chain,” “tropomyosin alpha-4 chain isoform X1,” “myosin-7-like and troponin T, slow skeletal muscle isoform X2” (Supplementary Table 2, Sheet E: “MostDiffExpressed BlackBand”). Other unigenes, within the 20 most differentially expressed unigenes in the black band integument transcriptome, correspond to “calmodulins-like,” “kelch-like protein,” “immunoglobulin superfamily member 22,” “homeobox protein BarH-like 2,” “keratin—type 1 cytoskeletal 18-like isoform X1,” “keratin—type II cytoskeletal 8-like” and “profilin-2-like isoform X3” (Supplementary Table 2, Sheet E: “MostDiffExpressed BlackBand”).
Enzymatic Diversity Evaluation Using PRIAM Annotation
Based on BLAST analyses performed using the PRIAM database as a reference, enzyme annotation was performed and allowed to identify transcripts coding for 346 oxidoreductases (Class EC 1.), 1,296 transferases (Class EC 2.), 937 hydrolases (Class EC 3.), 111 lyases (EC 4.), 101 isomerases (Class EC 5.), 163 ligases (Class EC 6.), and 0 translocases (Class EC 7.) for a total of 2,954 enzyme-coding transcripts for the general reference transcriptome (merged transcriptome) (Supplementary Table 3). On the 2,954 predicted enzyme-coding unigenes, 30 were uniquely found in the photogenic ventral skin transcriptome (i.e., absent from the black band transcriptome): 3 oxidoreductases, 14 transferases, 11 hydrolases, 1 lyase, and 1 isomerase. The same analysis was performed on the recently published ventral integument transcriptome of the lanternshark Etmopterus spinax (Delroisse et al., 2018) and allowed to identify a similar enzyme diversity: 302 oxidoreductases (Class EC 1.), 1,334 transferases (Class EC 2.), 876 hydrolases (Class EC 3.), 109 lyases (EC 4.), 95 isomerases (Class EC 5.), 149 ligases (Class EC 6.), and 0 translocases (Class EC 7.) for a total of 2,865 enzyme-coding transcripts.
The Search of Transcripts Coding for Luciferase-Like Candidates
BLAST analyses were used to identify transcripts coding for proteins similar to bacteria-type luciferases, insect-type luciferases (also characterizing the luminous cephalopod Watasenia and the Porifera Suberites) and to symplectin, a photoprotein isolated from the cephalopod Sthenoteuthis oulaniensis (Fujii et al., 2002). However reciprocal BLAST analyses revealed that these transcripts have more similarities to other related proteins, respectively, a «basic proline-rich protein-like», an «acyl-CoA synthetase family member 2, mitochondrial» and a «biotinidase isoform X1» (Supplementary Table 1). Only the “acyl-CoA synthetase family member 2, mitochondrial” appears to have a higher FPKM expression value in the ventral skin integument transcriptome. However, it is known that this metabolic enzyme is present in all multicellular organisms (Inouye, 2010) and that it was specifically co-opted into a luciferase in the insect lineages, in cephalopods and possibly in sponges (Müller et al., 2009; Gimenez et al., 2016; Delroisse et al., 2017b). These analyses support the hypothesis that I. brasiliensis is using a non-described biochemical system based on an unknown luciferase/photoprotein system.
Discussion
Deep-sea sharks exhibit various luminous patterns in terms of shape and color. As previously observed (Bennett, 1840; Widder, 1998), I. brasiliensis is shown to emit light ventrally, except at the level of the highly pigmented black band area. The bioluminescence color range of multiple deep-sea sharks is depicted in the literature (for review in Herring, 1983; Claes and Mallefet, 2009; Renwart et al., 2014; Claes et al., 2015; Duchatelet et al., 2019a): e.g., Etmopterus spinax, λmax = 486 nm; E. molleri, λmax = 477 nm; E. splendidus, λmax = 476 nm; Squaliolus aliae, λmax = 457 nm; Isistius brasiliensis, λmax = 455 nm. Although the cookie cutter shark I. brasiliensis was sometimes depicted as a greenish light emitter (Widder, 1998), our pictures of living specimens confirm a blue luminescence consistent with the maximum wavelength of emission measured (i.e., 455 nm).
The present study highlights that photophores are almost absent (i.e., very few residuals photophores were observed) from the cookie cutter shark black band area as suggested by Widder (1998). In parallel, this study presents the first analysis of the photophore morphology and coverage in I. brasiliensis (and, more generally, in Dalatiidae). As recently observed in other luminous sharks (Claes and Mallefet, 2009; Claes et al., 2015; Duchatelet et al., 2020b; Mallefet et al., 2021), I. brasiliensis photophores, visible as black dots between placoid scales, are mainly distributed within the ventral epidermis with a clear dorso-ventral gradient. Counterillumination (i.e., ventral light emission that mimic the residual downwelling light in terms of intensity, wavelength and angular distribution to avoid being spotted by underneath swimming predators; Clarke, 1963; Johnsen et al., 2004), as an anti-predatory tool, was suggested to be the bioluminescence function for Dalatiidae species (Widder, 1998, Claes et al., 2015). Our results suggest that, conversely to the Etmopteridae luminous pattern (Claes and Mallefet, 2008; Claes et al., 2010, 2013, 2015; Duchatelet et al., 2019b), the cookie-cutter shark did not show any specific pattern in the sexual areas, the flank marking, or the spine-free dorsal fins which support the hypothesis of a potentially unique counterillumination function of the light production in this species. Some authors also assumed that I. brasiliensis might use its non-luminescent black band, which looks as a small fish or crustacean in a downwelling light context where the remaining body part is hidden by counterillumination, to lure larger predator (such as bigger pelagic fish, cetaceans, sharks and seals) and to steal a piece of flesh from them via its adapted jaw (Widder, 1998).
Our analyses revealed a morphological organization of the photophore very similar to that observed in the genus Squaliolus (Dalatiidae, Seigel, 1978) and Zameus (Somniosidae, Duchatelet et al., 2021): a unique photocyte embedded in a pigmented sheath and surmounted by a small iris-like structure and lens cells. Therefore, an evolutive conservation of the light organ morphology is suggested in the families Dalatiidae and Somniosidae. Comparatively, the Etmopteridae shark family displays a more complex organization with multiple photocytes per photophores (Claes and Mallefet, 2009; Renwart et al., 2014; Claes et al., 2015; Duchatelet et al., 2019a).
In parallel, this study presents a first release of transcriptomic data for the species. Two transcriptomes were generated from integument fragments obtained from the black band region and the ventral skin region of one individual of I. brasiliensis. Several decades of research focused on more accessible shallow water-sharks, and genomic surveys and genetic data are available for only a few species. The complete genomes of Callorhinchus milii (Venkatesh et al., 2014), Rhincodon typus (Read et al., 2017), Chiloscyllium punctatum and Scyliorhinus torazame (Hara et al., 2018) enhanced the research on the origin of Gnathostomes and Vertebrates. The first transcriptome dataset of a deep-sea shark (Etmopterus spinax) has only recently been published (Delroisse et al., 2018) which highlight a lack of genomic/transcriptomic data for these deep-sea sharks. To our knowledge, the present study is the first characterization of a transcriptome of I. brasiliensis. Transcriptome annotation allowed to generate of a list of predicted transcripts expressed in the black band and the ventral integument regions. These datasets could contribute to the comprehension of the gene expression of this organism and possibly guide future research toward a better understanding of the biology of this deep-sea species.
Luciferases and photoproteins are usually not conserved among luminous animals. While all these enzymes perform the same biochemical task, yet with various molecular substrates, non-homologous luciferases and photoproteins emerged convergently in several metazoan lineages. As an example, while insects share a common type of luciferase used for their bioluminescence (Viviani et al., 2013), three other non-homologous types of luciferase have been described in crustaceans up to now (i.e., Cypridina/Vargula-type luciferase, Copepoda-type luciferase, and Oplophorus-type luciferases) (Inouye et al., 2000; Inouye and Sasaki, 2007). In some cases, however, a homologous system appears to be used in phylogenetically distant luminous organisms. Such a co-option event has been strongly suggested in echinoderms (Amphiura filiformis) and urochordates (Pyrosoma atlanticum) which most likely use a luciferase homologous to the one initially described in Cnidaria (Delroisse et al., 2017b; Tessler et al., 2020). In the cephalopod Watasenia, a luciferase homologous to the insect-type is used (Gimenez et al., 2016). For this reason, it is meaningful to search for luciferase candidates homologous to known luciferases in bioluminescent species for which the luminous system is undescribed. However, it has to be stated that a “new system,” i.e., based on a new type of enzyme not described yet, would not be detected by this approach. This situation is observed in the well-known ostracod luciferase (Cypridina noctiluca) and the fireworm luciferase (e.g., Odontosyllis undecimdonta) (Thompson et al., 1989; Nakajima et al., 2004; Mitani et al., 2018, 2019). Both types of luciferase indeed have no homology with known enzymes.
Luminous sharks appear as a remaining enigma because luminous compounds (i.e., luciferin and luciferase) are still unknown (Renwart and Mallefet, 2013). In the present study, we searched for sequences homologous to known luciferases or photoproteins in the transcriptome of I. brasiliensis. We then compared the relative expression of candidate transcripts between the photogenic ventral integument and the non-photogenic black band integument region. Because no specific expression profile was highlighted, our results suggest that I. brasiliensis uses a new biochemical system based on an undescribed luciferase/photoprotein system. This preliminary hypothesis, however, remains to be confirmed. Indeed, because of the presence of a very limited number of photophores within the black band, a strict absence of luciferase/photoprotein mRNA is unlikely in this tissue. In addition, luciferases are classically pleiotropic proteins that may have various biochemical activities and, for that reason, may be expressed in a large variety of tissues, including non-luminous ones (e.g., Fortova et al., 2013; Delroisse et al., 2017a).
As another way to following our hypothesis that I. brasiliensis might use an undescribed biochemical luminous system, we investigated the diversity of enzymes expressed within the integument of the tested individual. Several enzyme-coding transcripts related to metabolic processes such as glycolysis were shown to be highly expressed and differentially expressed within the transcriptomes. From a technical point of view, it is particularly challenging to separate skin layers from the rest of the integument without affecting the quality of the sample (i.e., quality of extracted mRNA). For this reason, it is difficult to avoid the presence underlying tissues (i.e., connective tissue and underlying muscles). In addition, the RNA extractions are generally considered as efficient for muscle tissues mainly due to the high transcript expression which imply that small difference in terms of muscular tissue mass might lead to strong upper or under representation of muscle-related transcripts. This can be seen as a limitation of our dataset. However, 30 enzyme-coding unigenes were predicted to be exclusively expressed in the photogenic ventral skin transcriptome and are apparently absent from the black band transcriptome. These 30 candidates could be the primary target of future investigations aiming to identify the luciferase of I. brasiliensis. The generated datasets therefore constitute interesting references that should be used, in the near future, to investigate the functional characterization of the integument of this deep-sea shark.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR12351835; https://www.ncbi.nlm.nih.gov/, SRR12351836. The refence assembled transcriptome is also available at: https://10.0.51.84/RG.2.2.12473.29288.
Ethics Statement
Animal procedures were conducted in compliance with Belgian national guidelines and in agreement with European directive 2010/63/UE, under the approval of the Animal Ethics Committee of the University of Louvain in Louvain-la-Neuve (Belgium). Living cookie-cutter shark from Australia was maintained and euthanized according to Museums Victoria animal care protocol.
Author Contributions
JD and LD performed classical histology, transcriptomic analyses, and wrote the first draft of the manuscript. JM collected the tissues and performed high sensitivity photography on living specimens. PF and JM supervised the work. All authors read, modified, and approved the final manuscript.
Funding
This work was supported by an FRS–FNRS Grant (T.0169.20) awarded to the Marine Biology Laboratory of the Catholic University of Louvain (UCLouvain) and the Biology of Marine Organisms and Biomimetics Unit of the University of Mons (UMONS).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Sampling the Abyss was led by Museums Victoria under Chief Scientist Tim O’Hara and supported by CSIRO Marine National Facility and NESP Marine Biodiversity Hub. We greatly thanks for the opportunity to collect Isistius brasiliensis specimens. We would also like to thank the reviewers for their thoughtful comments. JD, JM, and PF were, respectively, Postdoctoral Fellow, Research Associate, and Research Director of the Fund for Scientific Research of Belgium (F.R.S-FNRS). LD was postdoctoral researcher at the University of Louvain. This study was a contribution from the “Centre Interuniversitaire de Biologie Marine” (CIBIM).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2021.627045/full#supplementary-material
Supplementary Figure 1 | Photophore densities with the skin of I. brasiliensis.
Supplementary Figure 2 | Read quality evaluation. The graph shows the quality score (Phred score) across all bases for the clean reads obtained via Illumina X Ten sequencing. The box plot whiskers represent the quantile range (between the 10 and 90% quantiles). The yellow boxes represent the inter-quartile range (between the 25 and 75% quartiles). The red line is the median value. The blue line represents the mean quality.
Supplementary Table 1 | Luciferase reference sequences and BLAST/reciprocal BLAST analyses of the reference transcriptome of I. brasiliensis.
Supplementary Table 2 | Global transcriptome annotation (NR, NT, GO, COG, KEGG). Sheets (A–E) (A) unigene annotation, (B) 20 most expressed unigenes within the ventral transcriptome, (C) 20 most expressed unigenes within the black band transcriptome, (D) 20 most differentially expressed unigenes within the ventral transcriptome, (E) 20 most differentially expressed unigenes within the black band transcriptome.
Supplementary Table 3 | Enzyme repertoire annotation based on the PRIAM database.
Footnotes
- ^ www.mnf.csiro.au/en/Voyages/IN2017_V03
- ^ www.geneontology.org/
- ^ www.fa.cgu.edu.tw
- ^ www.usegalaxy.eu
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed November, 2020).
Audic, S., and Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Bennett, F. D. (1840). Narrative of a Whaling Voyage Round the Globe, from the Year 1833 to 1836, Vol. 2. London: Richard Bentley, 257–258.
Carrier, J. F., Musick, J. A., and Heithaus, M. R. (eds). (2012). Biology of Sharks and their relatives. CRC Marine Biology Series. Boca Raton, FL: CRC press.
Claes, J. M., Aksnes, D. L., and Mallefet, J. (2010). Phantom hunter of the fjords: camouflage by counterillumination in a shark (Etmopterus spinax). J. Exp. Mar. Biol. Ecol. 388, 28–32. doi: 10.1016/j.jembe.2010.03.009
Claes, J. M., and Mallefet, J. (2008). Early development of bioluminescence suggests camouflage by counter-illumination in the velvet belly lantern shark Etmopterus spinax (Squaloidea: Etmopteridae). J. Fish Biol. 73, 1337–1350. doi: 10.1111/j.1095-8649.2008.02006.x
Claes, J. M., and Mallefet, J. (2009). “Bioluminescence of sharks: first synthesis,” in Bioluminescence in Focus – A Collection of Illuminating Essays, ed. V. B. Meyer-Rochow (Kerala: Research Signpost), 51–65.
Claes, J. M., Dean, M. N., Nilsson, D.-E., Hart, N. S., and Mallefet, J. (2013). A deepwater fish with ‘lightsabers’ – dorsal spine-associated luminescence in a counterilluminating lanternshark. Sci. Rep. 3:1308. doi: 10.1038/srep01308
Claes, J. M., Ho, H.-C., and Mallefet, J. (2011). Control of luminescence from pygmy shark (Squaliolus aliae) photophores. J. Exp. Biol. 215, 1691–1699. doi: 10.1242/jeb.066704
Claes, J. M., Nilsson, D.-E., Straube, N., Collin, S. P., and Mallefet, J. (2015). Iso-luminance counterillumination drove bioluminescent shark radiation. Sci. Rep. 4, 4328. doi: 10.1038/srep04328
Clarke, W. (1963). Function of bioluminescence in mesopelagic organisms. Nature 198, 1244–1246. doi: 10.1038/1981244a0
Cotton, C. F., and Grubbs, R. D. (2015). Biology of deep-water chondrichthyans: introduction. Deep Sea Res. 2 Top. Stud. Oceanogr. 115, 1–10. doi: 10.1016/j.dsr2.2015.02.030
Das, S., Shyamal, S., and Durica, D. S. (2016). Analysis of annotation and differential expression methods used in RNA-Seq studies in crustacean systems. Integr. Comp. Biol. 56, 1067–1079. doi: 10.1093/icb/icw117
Delroisse, J., Duchatelet, L., Flammang, P., and Mallefet, J. (2018). De novo transcriptome analyses provide insights into opsin-based photoreception in the lanternshark Etmopterus spinax. PLoS One 13:e0209767. doi: 10.1371/journal.pone.0209767
Delroisse, J., Flammang, P., and Mallefet, J. (2014). Marine luciferases: are they really taxon-specific? A putative luciferase evolved by co-option in an echinoderm lineage. Luminescence 29:15.
Delroisse, J., Mallefet, J., and Flammang, P. (2016). De novo adult transcriptomes of two European brittle stars: spotlight on opsin-based photoreception. PLoS One 11:e0152988. doi: 10.1371/journal.pone.0152988
Delroisse, J., Ortega-Martinez, O., Dupont, S., Mallefet, J., and Flammang, P. (2015). De novo transcriptome of the European brittle star Amphiura filiformis pluteus larvae. Mar. Genomics 23, 109–121. doi: 10.1016/j.margen.2015.05.014
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Eeckhaut, I., Flammang, P., and Mallefet, J. (2017a). Fine structure of the luminous spines and luciferase detection in the brittle star Amphiura filiformis. Zool. Anz. 269, 1–12. doi: 10.1016/j.jcz.2017.05.001
Delroisse, J., Ullrich-Lüter, E., Blaue, S., Ortega-Martinez, O., Eeckhaut, I., Flammang, P., et al. (2017b). A puzzling homology: a brittle star using a putative cnidarian-type luciferase for bioluminescence. Open Biol. 7:160300. doi: 10.1098/rsob.160300
Duchatelet, L., Delroisse, J., Flammang, P., Mahillon, J., and Mallefet, J. (2019a). Etmopterus spinax, the velvet belly lanternshark, does not use bacterial luminescence. Acta Histochem. 121, 516–521. doi: 10.1016/j.acthis.2019.04.010
Duchatelet, L., Delroisse, J., Pinte, N., Sato, K., Ho, H.-C., and Mallefet, J. (2020a). Adrenocorticotropic hormone and cyclic adenosine monophosphate are involved in the control of shark bioluminescence. Photochem. Photobiol. 96, 37–45. doi: 10.1111/php.13154
Duchatelet, L., Marion, R., and Mallefet, J. (2021). A third luminous shark family: confirmation of luminescence ability for Zameus squamulosus (Squaliformes; Somniosidae). Photochem. Photobiol. doi: 10.1111/php.13393 [Epub ahead of print].
Duchatelet, L., Pinte, N., Tomita, T., Sato, K., and Mallefet, J. (2019b). Etmopteridae bioluminescence: dorsal pattern specificity and aposematic use. Zool. Lett. 5:9. doi: 10.1186/s40851-019-0126-2
Duchatelet, L., Sugihara, T., Delroisse, J., Koyanagi, M., Rezsohazy, R., Terakita, A., et al. (2020b). From extraocular photoreception to pigment movement regulation: a new control mechanism of the lanternshark luminescence. Sci. Rep. 10:10195. doi: 10.1038/s41598-020-67287-w
Dwyer, S. L., and Visser, I. N. (2011). Cookie cutter shark (Isistius sp.) bites on cetaceans, with particular reference to killer whales (Orca) (Orcinus orca). Aquat. Mamm. 37, 111–138. doi: 10.1578/AM.37.2.2011.111
Ebert, D. A., Fowler, S. L., and Compagno, L. J. (2013). Sharks of the World: A Fully Illustrated Guide. Plymouth: Wild Nature Press.
Feunteun, A., de Schrevel, C., Verhaegen, M., Chevallier, D., Duchemin, M., Ziani, N., et al. (2018). First evaluation of the cookie cutter sharks (Isistius sp.) predation pattern on different cetacean species in Martinique. Environ. Biol. Fish. 101, 749–759. doi: 10.1007/s10641-018-0735-1
Fortova, A., Sebestova, E., Stepankova, V., Koudelakova, T., Palkova, L., Damborsky, J., et al. (2013). DspA from Strongylocentrotus purpuratus: the first biochemically characterized haloalkane dehalogenase of non-microbial origin. Biochimie 95, 2091–2096. doi: 10.1016/j.biochi.2013.07.025
Fujii, T., Ahn, J. Y., Kuse, M., Mori, H., Matsuda, T., and Isobe, M. (2002). A novel photoprotein from oceanic squid (Symplectoteuthis oualaniensis) with sequence similarity to mammalian carbon–nitrogen hydrolase domains. Biochem. Biophys. Res. Commun. 293, 874–879. doi: 10.1016/S0006-291X(02)00296-6
Gimenez, G., Metcalf, P., Paterson, N. G., and Sharpe, M. L. (2016). Mass spectrometry analysis and transcriptome sequencing reveal glowing squid crystal proteins are in the same superfamily as firefly luciferase. Sci. Rep. 6:27638. doi: 10.1038/srep27638
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Hara, Y., Yamaguchi, K., Onimaru, K., Kadota, M., Koyanagi, M., Keeley, S. D., et al. (2018). Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2, 1761–1771. doi: 10.1038/s41559-018-0673-5
Hart, N. S., and Collin, S. P. (2014). Sharks senses and shark repellents. Int. Zool. 10, 38–64. doi: 10.1111/1749-4877.12095
Herring, P. J. (1983). The spectral characteristics of luminous marine organisms. Proc. R. Soc. Lond. B. 220, 183–217. doi: 10.1098/rspb.1983.0095
Hoyos-Padilla, M., Papastamatiou, Y. P., O’Sullivan, J., and Lowe, C. G. (2013). Observation of an attack by a Cookiecutter shark (Isistius brasiliensis) on a white shark (Carcharodon carcharias). Pac. Sci. 67, 129–134. doi: 10.2984/67.1.10
Inouye, S. (2010). Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell. Mol. Life Sci. 67, 387–404. doi: 10.1007/s00018-009-0170-8
Inouye, S., and Sasaki, S. (2007). Overexpression, purification and characterization of the catalytic component of Oplophorus luciferase in the deep-sea shrimp, Oplophorus gracilirostris. Protein Exp. Purif. 56, 261–268. doi: 10.1016/j.pep.2007.08.002
Inouye, S., Watanabe, K., Nakamura, H., and Shimomura, O. (2000). Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase. FEBS Lett. 481, 19–25. doi: 10.1016/S0014-5793(00)01963-3
Jahn, A. E., and Haedrich, R. L. (1987). Notes on the pelagic squaloid shark Isistius brasiliensis. Biol. Oceanogr. 5, 297–309. doi: 10.1080/01965581.1987.10749519
Johnsen, S., Widder, E. A., and Mobley, C. D. (2004). Propagation and perception of bioluminescence: factors affecting counterillumination as a cryptic strategy. Biol. Bull. 207, 1–16. doi: 10.2307/1543624
Jones, E. C. (1971). Isistius brasiliensis, a squaloid shark, the probable cause of crater wounds on fishes and cetaceans. Fish. Bull. USA 69, 791–798.
Kyne, P. M., and Simpfendorfer, C. A. (2007). A Collation and Summarization of Available Data on Deepwater Chondrichthyans: Biodiversity, Life History and Fisheries. IUCN Shark Specialist Group. Available online at: https://www.researchgate.net/profile/Colin-Simpfendorfer/publication/228826523_A_collation_and_summarization_of_available_data_on_deepwater_chondrichthyans_Biodiversity_life_history_and_fisheries/links/0912f50aa7896a865d000000/A-collation-and-summarization-of-available-data-on-deepwater-chondrichthyans-Biodiversity-life-history-and-fisheries.pdf (accessed November, 2020).
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357. doi: 10.1038/nmeth.1923
Le Boeuf, B. J., McCosker, J. E., and Hewitt, J. (1987). Crater wounds on northern elephant seals: the Cookiecutter shark strikes again. Fish. Bull. 85, 387–392.
Li, B., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Lowe, C., and Goodman-Lowe, G. (1996). Suntanning in hammerhead sharks. Nature 383, 677–677. doi: 10.1038/383677a0
Mallefet, J., Stevens, D. W., and Duchatelet, L. (2021). Bioluminescence of the largest luminous vertebrate, the kitefin shark, Dalatias licha: first insights and comparative aspects. Front. Mar. Sci. 8:633582. doi: 10.3389/fmars.2021.633582
Meyer, W., and Seegers, U. (2012). Basics of skin structure and function in elasmobranchs: a review. J. Fish Biol. 80, 1940–1967. doi: 10.1111/j.1095-8649.2011.03207.x
Mitani, Y., Yasuno, R., Futahashi, R., Oakley, T. H., and Ohmiya, Y. (2019). Luciferase gene of a Caribbean fireworm (Syllidae) from Puerto Rico. Sci. Rep. 9:13015.
Mitani, Y., Yasuno, R., Isaka, M., Mitsuda, N., Futahashi, R., Kamagata, Y., et al. (2018). Novel gene encoding a unique luciferase from the fireworm Odontsyllis undecimdonta. Sci. Rep. 8, 12789.
Moore, K. S., Wehrli, S., Roder, H., Rogers, M., Forrest, J. N. Jr., McCrimmon, D., et al. (1993). Squalamine: an aminosterol antibiotic from the shark. Proc. Natl. Acad. Sci. U.S.A. 90, 1354–1358. doi: 10.1073/pnas.90.4.1354
Müller, W. E. G., Kasueske, M., Wang, X., Schröder, H. C., Wang, Y., Pisignano, D., et al. (2009). Luciferase a light source for the silica-based optical waveguides (spicules) in the demosponge Suberites domuncula. Cell. Mol. Life Sci. 66:537. doi: 10.1007/s00018-008-8492-5
Muñoz-Chápuli, R., Rel Salgado, J. C., and De La Serna, J. M. (1988). Biogeography of Isistius brasiliensis in the North-Eastern Atlantic, inferred from crater wounds on Swordfish (Xiphias gladius). J. Mar. Biol. Assoc. U. K. 68, 315–321. doi: 10.1017/S0025315400052218
Murakami, C., Yoshida, H., and Yonezaki, S. (2018). Cookie-cutter shark Isistius brasiliensis eats Bryde’s whale Balaenoptera brydei. Ichthyol. Res. 65, 398–404. doi: 10.1007/s10228-018-0619-6
Nakajima, Y., Kobayashi, K., Yamagishi, K., Enomoto, T., and Ohmiya, Y. (2004). cDNA cloning and characterization of a secreted luciferase from the luminous Japanese ostracod, Cypridina noctiluca. Biosci. Biotechnol. Biochem. 68, 565–570. doi: 10.1271/bbb.68.565
Oeffner, J., and Lauder, G. V. (2012). The hydrodynamic function of shark skin and two biomimetic applications. J. Exp. Biol. 215, 785–795. doi: 10.1242/jeb.063040
Papastamatiou, Y. P., Wetherbee, B. M., O’Sullivan, J., Goodmanlowe, G. D., and And Lowe, C. G. (2010). Foraging ecology of cookiecutter sharks (Isistius brasiliensis) on pelagic fishes in Hawaii, inferred from prey bite wounds. Environ. Biol. Fish. 88, 361–368. doi: 10.1007/s10641-010-9649-2
Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., et al. (2003). TIGR Gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652. doi: 10.1093/bioinformatics/btg034
Pollerspöck, J., and Straube, N. (2019). Bibliography Database of Living/Fossil Sharks, Rays and Chimaeras (Chondrichthyes: Elasmobranchii, Holocephali) – Papers of the year 2018, version 01/2019. World Wide Web electronic publication. Available online at: www.shark-references.com (accessed December, 2020).
Read, T. D., Petit, R. A., Joseph, S. J., Alam, M. T., Weil, M. R., Ahmad, M., et al. (2017). Draft sequencing and assembly of the genome of the world’s largest fish, the whale shark: Rhincodon typus smith 1828. BMC Genom. 18:532. doi: 10.1186/s12864-017-3926-9
Reif, W.-E. (1985). Functions of scales and photophores in mesopelagic luminescent sharks. Acta Zool. 66, 111–118. doi: 10.1111/j.1463-6395.1985.tb00829.x
Renwart, M., and Mallefet, J. (2013). First study of the chemistry of the luminous system in a deep-sea shark, Etmopterus spinax Linnaeus, 1758 (Chondrichthyes: Etmopteridae). J. Exp. Mar. Biol. Ecol. 448, 214–219. doi: 10.1016/j.jembe.2013.07.010
Renwart, M., Delroisse, J., Claes, J. M., and Mallefet, J. (2014). Ultrastructure organization of lantern shark (Etmopterus spinax, Linnaeus, 1758) photophores. Zoomorphology 133, 405–416. doi: 10.1007/s00435-014-0230-y
Renwart, M., Delroisse, J., Flammang, P., Claes, J. M., and Mallefet, J. (2015). Cytological changes during luminescence production in lanternshark (Etmopterus spinax Linnaeus, 1758) photophores. Zoomorphology 134, 107–116. doi: 10.1007/s00435-014-0235-6
Robbins, R., and Fox, A. (2012). Further evidence of pigmentation change in white sharks, Carcharodon carcharias. Mar. Freshw. Res. 63, 1215–1217. doi: 10.1071/MF12208
Seigel, J. A. (1978). Revision of the Dalatiid shark genus Squaliolus: anatomy, systematics, ecology. Copeia 1978, 602–614. doi: 10.2307/1443686
Straube, N., Li, C., Claes, J. M., Corrigan, S., and Naylor, G. J. P. (2015). Molecular phylogeny of Squaliformes and first occurrence of bioluminescence in sharks. BMC Evol. Biol. 15:162. doi: 10.1186/s12862-015-0446-6
Tessler, M., Gaffney, J. P., Oliveira, A. G., Guarnaccia, A., Dobi, K. C., Gujarati, N. A., et al. (2020). A putative chordate luciferase from a cosmopolitan tunicate indicates convergent bioluminescence evolution across phyla. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-73446-w
Thompson, E. M., Nagata, S., and Tsuji, F. I. (1989). Cloning and expression of cDNA for the luciferase from the marine ostracod Vargula hilgendorfii. Proc. Natl. Acad. Sci. 86, 6567–6571. doi: 10.1073/pnas.86.17.6567
Tsutsui, S., Dotsuta, Y., Ono, A., Suzuki, M., Tateno, H., Hirabayashi, J., et al. (2015). A C-type lectin isolated from the skin of Japanese bullhead shark (Heterondontus japonicus) binds a remarkably broad range of sugars and induces blood coagulation. J. Biochem. 157, 345–356. doi: 10.1093/jb/mvu080
Venkatesh, B., Lee, A. P., Ravi, V., Maurya, A. K., Lian, M. M., Swann, J. B., et al. (2014). Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174–179. doi: 10.1038/nature12826
Visconti, M. A., Ramanzini, G. C., Camargo, C. R., and Castrucci, A. M. L. (1999). Elasmobranch color change: a short review and novel data on hormone regulation. J. Exp. Zool. 284, 485–491. doi: 10.1002/(SICI)1097-010X(19991001)284:5<485::AID-JEZ3<3.0.CO;2-5
Viviani, V. R., Prado, R. A., Neves, D. R., Kato, D., and Barbosa, J. A. (2013). A route from darkness to light: emergence and evolution of luciferase activity in AMP-CoA-ligases inferred from a mealworm luciferase-like enzyme. Biochemistry 52, 3963–3973. doi: 10.1021/bi400141u
Wainwright, S. A., Vosburgh, F., and Hebrank, J. H. (1978). Shark skin: function in locomotion. Science 202, 747–749. doi: 10.1126/science.202.4369.747
Watkins, O. C., Sharpe, M. L., Perry, N. B., and Krause, K. L. (2018). New Zealand glowworm (Arachnocampa luminosa) bioluminescence is produced by a firefly-like luciferase but an entirely new luciferin. Sci. Rep. 8:3278. doi: 10.1038/s41598-018-21298-w
Keywords: bioluminescent shark, cookie-cutter shark, skin, transcriptome, enzyme diversity
Citation: Delroisse J, Duchatelet L, Flammang P and Mallefet J (2021) Photophore Distribution and Enzymatic Diversity Within the Photogenic Integument of the Cookie-Cutter Shark Isistius brasiliensis (Chondrichthyes: Dalatiidae). Front. Mar. Sci. 8:627045. doi: 10.3389/fmars.2021.627045
Received: 07 November 2020; Accepted: 22 March 2021;
Published: 15 April 2021.
Edited by:
Mark S. Springer, University of California, Riverside, United StatesReviewed by:
Yasuo Mitani, National Institute of Advanced Industrial Science and Technology (AIST), JapanTakushi Kishida, Museum of Natural and Environmental History, Shizuoka, Japan
Robert William Meredith, Montclair State University, United States
Copyright © 2021 Delroisse, Duchatelet, Flammang and Mallefet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jérôme Delroisse, Jerome.Delroisse@umons.ac.be; Laurent Duchatelet, Laurent.Duchatelet@uclouvain.be
†These authors have contributed equally to this work
 Jérôme Delroisse
Jérôme Delroisse Laurent Duchatelet
Laurent Duchatelet Patrick Flammang
Patrick Flammang Jérôme Mallefet
Jérôme Mallefet