DNA Barcoding Technology Used to Successfully Sub-Classify a Museum Whale Specimen as Balaenoptera edeni edeni
- 1Institute of Archaeological Science, Fudan University, Shanghai, China
- 2State Key Laboratory of Estuarine and Coastal Research, East China Normal University, Shanghai, China
- 3China (Hainan) Museum of the South China Sea, Qionghai, China
- 4Ministry of Education Key Laboratory of Contemporary Anthropology, Department of Anthropology and Human Genetics, School of Life Sciences, Fudan University, Shanghai, China
DNA barcoding technology is becoming an increasingly powerful tool in resolving issues of detailed species identification based on morphology, as commonly employed by museums. In the present study, we aimed to identify a stranded Bryde’s whale on Hainan Island, China by extracting DNA from a vertebra pre-treated by physical and/or chemical processes. Based on morphological characteristics, this Bryde’s whale was initially determined as Balaenoptera edeni. Then, DNA was efficiently extracted using ancient DNA techniques. The mitochondrial gene (COI) phylogenetic analysis further revealed that this museum whale specimen belonged to the sub-species B. e. edeni. This study provides a testable and rapid method for museum species verification, by using ancient DNA extraction methods to compensate the disadvantage of traditional DNA extraction methods that are difficult to extract valid DNA.
Introduction
Identifying the species/sub-species of museum specimens has long been a major challenge. Traditional approaches to identification have been based on morphometric analysis and/or morphological criteria, often without the services of taxonomic specialists (Bacher, 2012). Genetic materials have recently emerged as a promising trend in the rapid resolution of species/sub-species identification for both fresh and ancient museum specimens (Barbanera et al., 2020; Pierson et al., 2020). These “non-invasive” approaches cost museums little to nothing with regard to the quantity and quality of specimens held. This development has been offset by the high degree of decomposition among much museum material, whose prior physical or chemical treatment can severely impede the process of effective DNA extraction. As such, increasingly effective means have been developed for the extraction of highly fragmented DNA in the presence of contaminants and inhibitors (Rohland et al., 2018). One specific new method, DNA barcoding, takes advantage of short standardized sequences in order to facilitate species identification (Hebert et al., 2003; Savolainen et al., 2005). In DNA barcoding, both intraspecific variation and interspecific divergence can be significant, with the mitochondrial cytochrome c oxidase 1 (COI) identified as the best gene based on its conserved amino acid sequence, and hence the key to distinguishing animal species and sub-species (Knowlton and Weigt, 1998; Hebert et al., 2003; Chapuis et al., 2016). The DNA barcoding approach has been used to identify a variety of museum species/sub-species ranging from insects to fishes to primates (Thomsen et al., 2009; Hawlitschek et al., 2017). Nevertheless, DNA barcoding of marine mammals—the subject of this study—remains in its relative infancy.
Stranded cetacean specimens are objects of public fascination when displayed in the exhibition halls of museums. Bryde’s whale, or Bryde’s whale complex, a baleen whale occupying warm-temperate waters on a year-round basis, can be recognized by the three distinct ridges on its rostrum (Penry et al., 2018). Bryde’s whale is currently recognized as a single species (Balaenoptera edeni Anderson, 1879) with two recognized subspecies: a small coastal form (Eden’s whale, B. e. edeni) and a large oceanic form (Bryde’s whale, B. e. brydei) (Constantine et al., 2018; Penry et al., 2018; Liu et al., 2021; Committee on Taxonomy, 2022). Recently, sightings of Bryde’s whale have been recorded from the coasts of East and Southeast Asia (Yamada et al., 2008; Chen et al., 2019; Liu et al., 2021), south west Indian Ocean (Penry et al., 2018), Southern Africa (Best, 2001; Penry et al., 2011) and Gulf of Mexico (Rosel and Wilcox, 2014). However, as the type specimen for B. e. brydei was not designated with the naming of the species, and genetic analysis of the type specimen of B. e. edeni was not completed, detailed taxonomy within the Bryde’s whale group is unclear (Anderson, 1879; Constantine et al., 2015).
Tracking back at least four decades (1978–2016), about nine Bryde’s-like whales were stranded along the coast of Hainan Province, China, whereas little information was available on age, gender and taxonomy (Zhang et al., 2015; Liu et al., 2019). In 2019, an adult Bryde’s-like whale was discovered along the coast of Qiaotou Town, Chengmai County, Hainan Province, China. The specimen’s corpse had already been buried by nearby villagers by the time the museum team arrived on site. Based on the extent of decomposition, the specimen was judged to have been deceased for approximately 2 weeks, and to have floated on the ocean for over a week prior to washing ashore. For better preservation, the carcass was subsequently transported to the museum. Whale skeleton, skin and residual tissue were repeatedly steamed at high temperatures, and finally soaked by chemical method using an anti-mold agent. Based on morphological characteristics (the presence of the diagnostic Bryde’s whales triple head ridge consisting of a central ridge flanked by two lateral rostral ridges; Yamada, 2009; Constantine et al., 2018), the specimen was evaluated as Bryde’s whale complex. However, no other features were available for further species/sub-species identification due to the high level of decomposition, leaving the sub-species undefined.
In the present study, we used ancient DNA methods and extracted DNA from the specimen’s vertebra. Considering that museum specimens are usually treated by physical and/or chemical processes, we expected to find a more suitable DNA extraction method, and to achieve a precise species/sub-species identification through DNA analysis. We therefore sequenced one fragment of the mitochondrial gene (COI) and constructed a phylogenetic tree on this basis. Our expectation was to identify the sub-species of this stranded Bryde’s whales by a DNA barcoding method outlined above.
Materials and Methods
To verify the sub-species, a sample of the specimen was extracted and its DNA sequenced for further analyses. A section of vertebra was selected and rinsed with distilled water. After drilling off some surface bone with a sterile drill bit, bone powder was then collected using a new sterile bit. According to precautions established by previously published ancient human DNA (Knapp et al., 2012; Xiong et al., 2022), genomic DNA was extracted in a dedicated aDNA facility at Fudan University. In total, 200 mg of bone power was used for DNA extraction (no sample power was used as negative control) by rotating overnight with 0.25 mg/ml Proteinase K (Merck, Germany) and 0.5 M EDTA (pH 8.0) at 37°C. After centrifuging, the supernatant was added to binding buffer [5 M GuHCl, 40% Isopropanol, 25 mM sodium acetate, and 0.05% Tween-20 (PH 5.2)] and magnetic beads (Enlighten Biotech, China). Then, DNA was eluted by TET buffer (QIAGEN, Germany). Finally, DNA concentration was quantified using a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, United States).
A∼700 bp segment of the mitochondrial COI gene was amplified to verify the specimen using primer pairs Balaenoptera-COI-F2 (ACACTAATCGGAGATGACCAAGTC) and Balaenoptera-COI-R2 (CTGATGTGAAATATGCTCGCG), designed by Primer Premier 5.0 (Lalitha, 2000). Polymerase chain reaction (PCR) was carried out in a total volume of 20 μL, consisting of 1 μL of genomic DNA, 1 μL of each primer, 7μL ddH2O, and 10 μL Premix Taq (TaKaRa, Japan). The PCR temperature profile was as follows: incubation at 94°C for 3 min, 14 cycles of 30 s at 94°C, 30 s at 62°C (–0.5°C every cycle), and 30 s at 72°C; then 20 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; and a final extension at 72°C for 10 min. PCR products were then purified and sequenced with forward primers on an ABI PRISM 3730 DNA capillary sequencer at MAP Tech (China). Newly obtained sequence with high quality was checked and submitted to GenBank under accession number: ON459534.
The COI nucleotide sequence was searched for its similarity using BLAST program from GenBank. Then, the relevant sequences were retrieved as reference sequences where available (mysticate families: Balaenopteridae, Eschrichtidea, Neobalaenidae, and Balaenide). The COI sequence was aligned with reference sequences using Clustal W (Thompson et al., 1994) in MEGA X (Kumar et al., 2018). The alignment was inspected visually and trimmed to the length of the shortest sequence. Phylogenetic analysis was performed on the alignment using the maximum-likelihood method in MEGA X with the bootstrap resampled 1,000 times. Here, a general time-reversible model with a gamma distribution (GTR + G) was gauged as the best-fit substitution model according to the corrected Akaike information criterion, using jModelTest v 2.1.3 (Darriba et al., 2012). Furthermore, the genetic distance (p-distance) of Balaenopteridae, Eschrichtiidae, Neobalaenidae and Balaenidae was calculated using MEGA X. To describe the intraspecific variation and relationship between newly obtained sequence and other related species (Balaenoptera edeni edeni, Balaenoptera edeni brydei, Balaenoptera borealis, and Balaenoptera omurai; these reference sequences were obtained from GenBank), a haplotype network was constructed by HAPLOVIEWER (Salzburger et al., 2011). Unique COI haplotypes were identified in DnaSP 6 (Rozas et al., 2017).
Results and Discussion
Other morphological features from the stranded whale specimen provided evidence for the presence of the Bryde’s whale complex (Figure 1A). The key features of the Bryde’s whale (three head ridges) were obvious macroscopically. The specimen’s body size, estimated to have reached around 12.5 m in length (Figure 1B), was much larger than previous specimens found in the South China Sea (Liu et al., 2021). After physical and/or chemical treatments, the complete skeleton was presented in the museum (Figure 1C).

Figure 1. Photographs of the museum whale specimen in this study. (A) Uncovering the stranded specimen; (B) whole body post-treatment, with characteristic three rostral cephalic ridges used as morphologic identification criterion; (C) complete skeleton on display in museum.
Previous research argued for similar morphological characteristics between B. e. brydei and B. e. edeni (Constantine et al., 2018; Penry et al., 2018; Castro et al., 2021). Generally, the body length of B. e. brydei may exceed B. e. edeni (Liu et al., 2021). Here, this stranded specimen is the largest individual of B. edeni recorded along the coast of Hainan Province and was initially considered as possibly B. e. brydei. Identifying the sub-species of the Bryde’s whale specimen in this study was further problematized by serious specimen decomposition. In order to extract DNA successfully, we employed extraction approaches for ancient DNA from a section of vertebrae, avoiding issues such as high temperature, degreasing and EtOH or formalin fixation that are known causes of DNA extraction failures (Ruane and Austin, 2017; McGuire et al., 2018; Pierson et al., 2020). The DNA concentration was to 0.854 ng/μl and suitable for the subsequent analysis. Additional sub-species confirmation was possible after the mitochondrial COI gene (722 bp) of our specimen was successfully sequenced. The phylogenetic tree based on COI (resulting in a 424 bp alignment) of four families (Balaenopteridae, Eschrichtidea, Neobalaenidae and Balaenide; 26 reference sequences provided in Supplementary Table 1) within the Mysticeti (baleen whale) group was then reconstructed (Figure 2). This phylogenetic tree was congruent with relationships derived from previous combined parsimony analysis of 23 datasets (including morphology, transposon insertions, mitochondrial genomes, cetacean satellite sequences and so on; see Gatesy et al. (2013). Moreover, the phylogenetic relationship for Bryde’s like, Sei, and Omura’s whales also revealed the same pattern as found previously, split into four clades, corresponding to B. e. edeni, B. e. brydei, B. borealis and B. omurai (Rosel et al., 2021). Newly obtained sequence from China belonged to the B. e. edeni clade. Based on genetic distance analysis, it showed that the genetic relationship is close between this sequence and B. e. edeni (0.000–0.002; see Supplementary Table 2). The haplotype network of Bryde’s like, Sei, and Omura’s whales consisted of 12 haplotypes (1 newly obtained sequence and 30 reference sequences shown in Figure 3; reference sequences in Supplementary Table 3). Specifically, according to geographical origin, this newly obtained sequence from China, belonged to the lineage of B. e. edeni, and shared a COI haplotype (515 bp) with B. e. edeni from Japan (AB201258 and NC_007938) and India (JN190945 and GQ856370). In our study, genetic analysis by DNA barcoding (COI) indicated that this Bryde’s whale belonged to B. e edeni, a result that could not be conclusively confirmed based on morphology alone. We have proved the efficacy of genetic analysis for identifying cetacean museum specimens to the sub-species level, especially for specimens with non-obvious morphological characteristics, requiring minimal sample sizes without conferring visible damage (Gilbert et al., 2007; Rowley et al., 2007). In our analysis, phylogenetic analysis and haplotype networks provided ample confirmation of species/sub-species identity. This study shows that ancient DNA techniques and DNA barcoding technology can compensate for lack of morphological identification, making it amenable to questions of species/sub-species identification in the museum context.
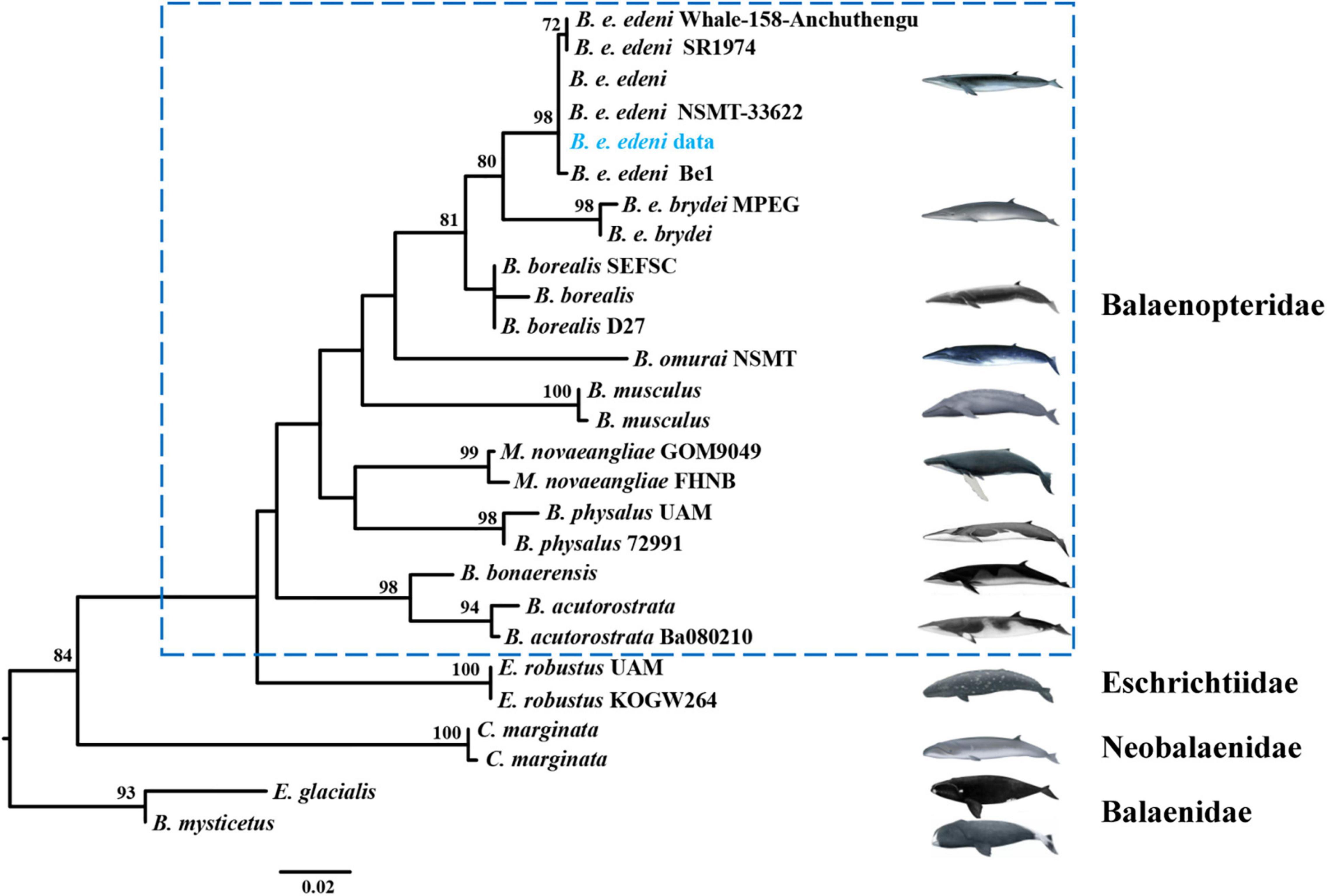
Figure 2. Phylogenetic tree showing the relationship among Mysticeti. Bootstrap support values shown on each node. Data on references sequences provided in Supplementary Table 1. Bootstrap support values of under 70% are not displayed. Newly obtained sequence from present study highlighted in blue font.
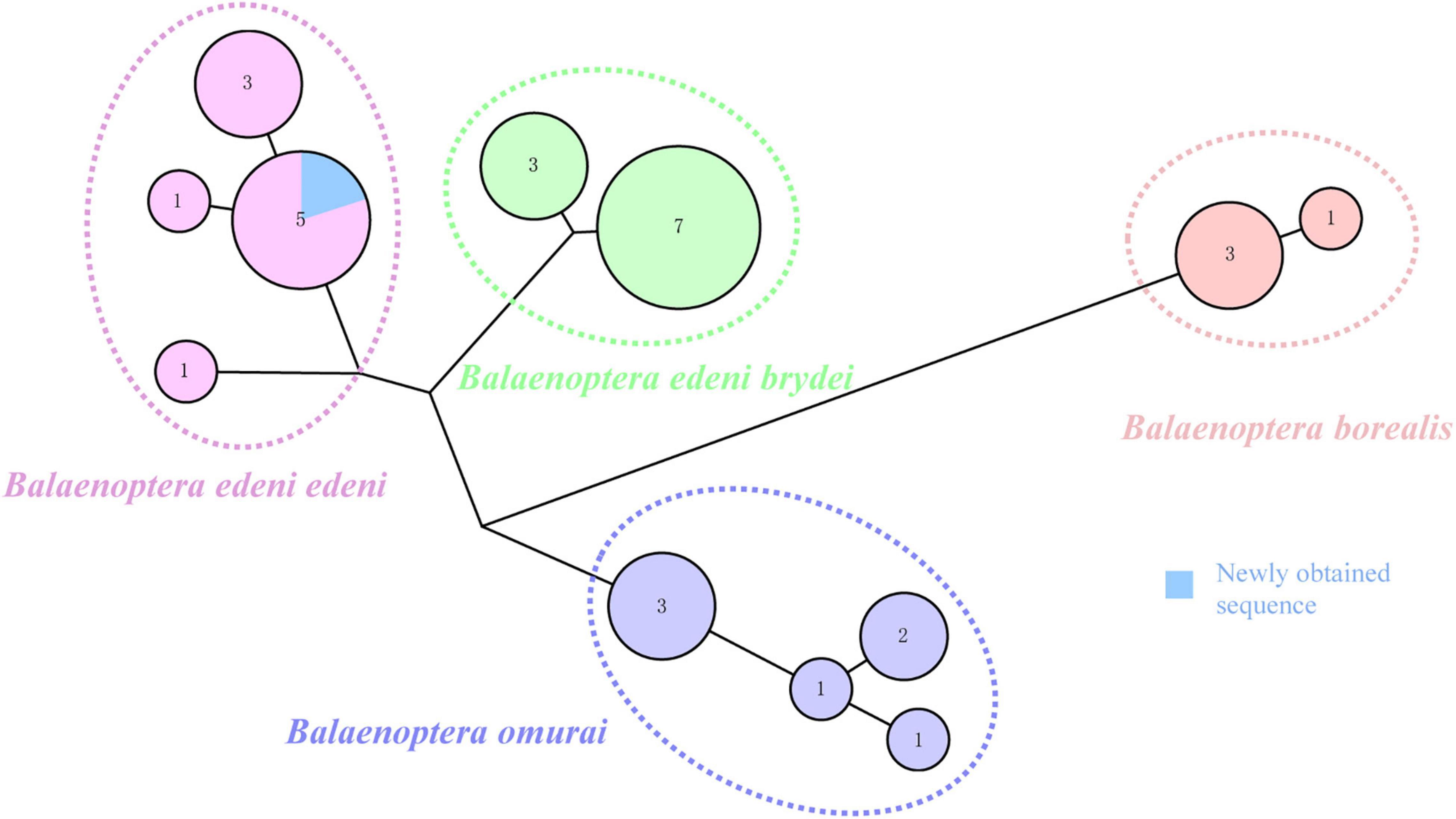
Figure 3. Haplotype network of Bryde’s like, Sei and Omura’s whales based on the COI gene (515 bp). Each circle represents a unique haplotype and its size indicates the number of individuals carrying the haplotype. Color coding allow easy discrimination of species in the complex. Blue coloring represents a newly obtained sequence. Data for haplotypes from GenBank provided in Supplementary Table 3.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/ON459534.
Ethics Statement
The animal study was reviewed and approved by Ethics Committee of Fudan University of Life Sciences.
Author Contributions
YF and SW designed and supervised the study. YF provided materials and resources. XR performed genetic laboratory work. XM performed genetic data analysis. EA integrated the genetic data. XR, XM, EA, YF, and SW wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (32070576) to SW and the National Natural Science Foundation of China (31900308) to XM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2022.921106/full#supplementary-material
Supplementary Table 1 | List of references sequences from Balaenopteridae, Eschrichtiidae, Neobalaenidae and Balaenidae in the COI phylogenetic analyses.
Supplementary Table 2 | The genetic distance (p-distance) of Balaenopteridae, Eschrichtiidae, Neobalaenidae and Balaenidae based on COI.
Supplementary Table 3 | List of reference sequences from GenBank used in the haplotype network analyses.
References
Anderson, J. (1879). Anatomical and Zoological Researches: Comprising an Account of the Zoological Results of the Two Expeditions to Western Yunnan in 1868 and 1875; and a Monograph of the Two Cetacean Genera, Platanista and Orcella. London: B. Quaritch, doi: 10.5962/bhl.title.50434
Bacher, S. (2012). Still not enough taxonomists: reply to Joppa et al. Trends Ecol. Evol. 27, 65–66. doi: 10.1016/j.tree.2011.11.003
Barbanera, F., Moretti, B., Guerrini, M., Al-Sheikhly, O. F., and Forcina, G. (2020). Investigation of ancient DNA to enhance natural history museum collections: misidentification of smooth-coated otter (Lutrogale perspicillata) specimens across multiple museums. Belg. J. Zool. 146, 101–112. doi: 10.26496/bjz.2016.45
Best, P. B. (2001). Distribution and population separation of Bryde’s whale Balaenoptera edeni off southern Africa. Mar. Ecol. Prog. Ser. 220, 277–289. doi: 10.3354/meps220277
Castro, J., Cid, A., and Laborde, M. I. (2021). Bryde’s whale (Balaenoptera edeni) new record for mainland Portugal. J. Cetacean Res. Manage. 22, 75–80. doi: 10.47536/jcrm.v22i1.333
Chapuis, M.-P., Bazelet, C. S., Blondin, L., Foucart, A., Vitalis, R., and Samways, M. J. (2016). Subspecific taxonomy of the desert locust, Schistocerca gregaria (Orthoptera: Acrididae), based on molecular and morphological characters. Syst. Entomol. 41, 516–530. doi: 10.1111/syen.12171
Chen, B., Zhu, L., Jefferson, T. A., Zhou, K., and Yang, G. (2019). Coastal Bryde’s Whales’ (Balaenoptera edeni) Foraging Area Near Weizhou Island in the Beibu Gulf. Aquat. Mamm. 45, 274–280. doi: 10.1578/AM.45.3.2019.274
Committee on Taxonomy (2022). List of Marine Mammal Species and Subspecies. Soc. Mar. Mammal. Available online at: http://marinemammalscience.org. (accessed May 2022).
Constantine, R., Iwata, T., Nieukirk, S. L., and Penry, G. S. (2018). Future Directions in Research on Bryde’s Whales. Front. Mar. Sci. 5:333. doi: 10.3389/fmars.2018.00333
Constantine, R., Johnson, M., Riekkola, L., Jervis, S., Kozmian-Ledward, L., Dennis, T., et al. (2015). Mitigation of vessel-strike mortality of endangered Bryde’s whales in the Hauraki Gulf, New Zealand. Biol. Conserv. 186, 149–157. doi: 10.1016/j.biocon.2015.03.008
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and high-performance computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Gatesy, J., Geisler, J. H., Chang, J., Buell, C., Berta, A., Meredith, R. W., et al. (2013). A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 66, 479–506. doi: 10.1016/j.ympev.2012.10.012
Gilbert, M. T. P., Moore, W., Melchior, L., and Worobey, M. (2007). DNA extraction from dry museum beetles without conferring external morphological damage. PLoS One 2:e272. doi: 10.1371/journal.pone.0000272
Hawlitschek, O., Toussaint, E. F. A., Gehring, P.-S., Ratsoavina, F. M., Cole, N., Crottini, A., et al. (2017). Gecko phylogeography in the Western Indian Ocean region: the oldest clade of Ebenavia inunguis lives on the youngest island. J. Biogeogr. 44, 409–420. doi: 10.1111/jbi.12912
Hebert, P. D. N., Cywinska, A., Ball, S. L., and deWaard, J. R. (2003). Biological identifications through DNA barcodes. Proc. Biol. Sci. 270, 313–321. doi: 10.1098/rspb.2002.2218
Knapp, M., Clarke, A. C., Horsburgh, K. A., and Matisoo-Smith, E. A. (2012). Setting the stage - building and working in an ancient DNA laboratory. Ann. Anat. Anat. Anz. 194, 3–6. doi: 10.1016/j.aanat.2011.03.008
Knowlton, N., and Weigt, L. A. (1998). New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. Lond. B Biol. Sci. 265, 2257–2263. doi: 10.1098/rspb.1998.0568
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lalitha, S. (2000). Primer Premier 5. Biotech. Softw. Internet Rep. 1, 270–272. doi: 10.1089/152791600459894
Liu, M., Lin, M., Zhang, P., Xue, T., and Li, S. (2019). An overview of cetacean stranding around Hainan Island in the South China Sea, 1978–2016: implications for research, conservation and management. Mar. Policy 101, 147–153. doi: 10.1016/j.marpol.2018.04.029
Liu, M., Lin, W., Lin, M., Liu, B., Dong, L., Zhang, P., et al. (2021). The First Attempt of Satellite Tracking on Occurrence and Migration of Bryde’s Whale (Balaenoptera edeni) in the Beibu Gulf. J. Mar. Sci. Eng. 9:796. doi: 10.3390/jmse9080796
McGuire, J. A., Cotoras, D. D., O’Connell, B., Lawalata, S. Z., Wang-Claypool, C. Y., Stubbs, A., et al. (2018). Squeezing water from a stone: high-throughput sequencing from a 145-year old holotype resolves (barely) a cryptic species problem in flying lizards. PeerJ 6:e4470. doi: 10.7717/peerj.4470
Penry, G. S., Hammond, P. S., Cockcroft, V. G., Best, P. B., Thornton, M., and Graves, J. A. (2018). Phylogenetic relationships in southern African Bryde’s whales inferred from mitochondrial DNA: further support for subspecies delineation between the two allopatric populations. Conserv. Genet. 19, 1349–1365. doi: 10.1007/s10592-018-1105-4
Penry, G., Cockcroft, V., and Hammond, P. (2011). Seasonal fluctuations in occurrence of inshore Bryde’s whales in Plettenberg Bay, South Africa, with notes on feeding and multispecies associations. Afr. J. Mar. Sci. 33, 403–414. doi: 10.2989/1814232x.2011.637617
Pierson, T. W., Kieran, T. J., Clause, A. G., and Castleberry, N. L. (2020). Preservation-Induced Morphological Change in Salamanders and Failed DNA Extraction from a Decades-Old Museum Specimen: Implications for Plethodon ainsworthi. J. Herpetol. 54:137. doi: 10.1670/19-012
Rohland, N., Glocke, I., Aximu-Petri, A., and Meyer, M. (2018). Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nat. Protoc. 13, 2447–2461. doi: 10.1038/s41596-018-0050-5
Rosel, P. E., and Wilcox, L. A. (2014). Genetic evidence reveals a unique lineage of Bryde’s whales in the northern Gulf of Mexico. Endanger. Species Res. 25, 19–34. doi: 10.3354/esr00606
Rosel, P. E., Wilcox, L. A., Yamada, T. K., and Mullin, K. D. (2021). A new species of baleen whale (Balaenoptera) from the Gulf of Mexico, with a review of its geographic distribution. Mar. Mamm. Sci. 37, 577–610. doi: 10.1111/mms.12776
Rowley, D. L., Coddington, J. A., Gates, M. W., Norrbom, A. L., Ochoa, R. A., Vandenberg, N. J., et al. (2007). Vouchering DNA-barcoded specimens: test of a nondestructive extraction protocol for terrestrial arthropods. Mol. Ecol. Notes 7, 915–924. doi: 10.1111/j.1471-8286.2007.01905.x
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., et al. (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 34, 3299–3302. doi: 10.1093/molbev/msx248
Ruane, S., and Austin, C. C. (2017). Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol. Ecol. Resour. 17, 1003–1008. doi: 10.1111/1755-0998.12655
Salzburger, W., Ewing, G. B., and Von Haeseler, A. (2011). The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol. Ecol. 20, 1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x
Savolainen, V., Cowan, R. S., Vogler, A. P., Roderick, G. K., and Lane, R. (2005). Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Philos. Trans. R. Soc. B Biol. Sci. 360, 1805–1811. doi: 10.1098/rstb.2005.1730
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Thomsen, P. F., Elias, S., Gilbert, M. T. P., Haile, J., Munch, K., Kuzmina, S., et al. (2009). Non-Destructive Sampling of Ancient Insect DNA. PLoS One 4:e5048. doi: 10.1371/journal.pone.0005048
Xiong, J., Du, P., Chen, G., Tao, Y., Zhou, B., Yang, Y., et al. (2022). Sex-Biased Population Admixture Mediated Subsistence Strategy Transition of Heishuiguo People in Han Dynasty Hexi Corridor. Front. Genet. 13:827277. doi: 10.3389/fgene.2022.827277
Yamada, T. K. (2009). “Omura’s Whale: Balaenoptera omurai,” in Encyclopedia of Marine Mammals (Second Edition), eds W. F. Perrin, B. Würsig, and J. G. M. Thewissen (London: Academic Press), 799–801. doi: 10.1016/B978-0-12-373553-9.00187-5
Yamada, T. K., Kakuda, T., and Tajima, Y. (2008). Middle sized balaenopterid whale specimens in the Philippines and Indonesia. Mem. Natl. Sci. Mus. Tokyo 45, 75–83.
Zhang, P., Li, S., Lin, M., and Xing, L. (2015). Database of Cetacean Stranding Records around Hainan Island Science Data Bank. Available Online at: http://doi.org/10.11922/sciencedb.37 (accessed on Jun 25, 2022).
Keywords: museum specimen, Bryde’s whale, morphological identification, DNA barcoding, DNA identification
Citation: Ren X, Ma X, Allen E, Fang Y and Wen S (2022) DNA Barcoding Technology Used to Successfully Sub-Classify a Museum Whale Specimen as Balaenoptera edeni edeni. Front. Ecol. Evol. 10:921106. doi: 10.3389/fevo.2022.921106
Received: 15 April 2022; Accepted: 13 June 2022;
Published: 14 July 2022.
Edited by:
Jonathan J. Fong, Lingnan University, ChinaReviewed by:
Utpal Smart, University of North Texas Health Science Center, United StatesSantiago Castroviejo-Fisher, Seville University, Spain
Copyright © 2022 Ren, Ma, Allen, Fang and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuan Fang, 751938749@qq.com; Shaoqing Wen, wenshaoqing@fudan.edu.cn
†These authors have contributed equally to this work
 Xiaoying Ren
Xiaoying Ren Xiaolin Ma2†
Xiaolin Ma2†  Edward Allen
Edward Allen Yuan Fang
Yuan Fang Shaoqing Wen
Shaoqing Wen