The genus Champia was established by Desvaux (1809) based on C. lumbricalis (Linnaeus) Desvaux, and is characterized by having an inner hollow thallus throughout the entire plant, cutting off a single two-celled auxiliary cell branch from a supporting cell, developing carposporangia only from the terminal cells of multicellular gonimoblast filaments, and having a fully elevated cystocarp with a prominent ostiole (Womersley 1996). Species within this genus have been recognized mainly by thallus shape and branching pattern rather than by reproductive structures (Park and Lee 1998).
Four species have been reported in Korea such as Champia compressa Harvey, C. expansa Yendo, C. japonica Okamura, and C. parvula (C. Agardh) Harvey (Lee and Kang 2001). Champia compressa was reported as C. bifida Okamura in Korea previously, but C. bifida was transferred to C. compressa based on a phenological study and morphological comparison of two type specimens (Park and Lee 1998). Park and Lee (1998) observed the thallus structure of gametophytes and tetrasporophytes of Champia from Korea. They discriminated four Champia species based on morphological features, such as a terete (C. parvula), compressed (C. compressa and C. japonica) or flattened (C. expansa) hollow thallus with segments separated by monostromatic septa or diaphragms, and multiaxial construction with longitudinal filaments lining the inner cortex. Thalli are branched irregularly (C. japonica, C. parvula), oppositely (C. compressa) or dichotomously (C. expansa) (Okamura 1901, Yendo 1903, Womersley 1996, Park and Lee 1998, Lozada-Troche and Ballantine 2010).
Champia compressa was established by Harvey (1838) based on materials collected from Muizenberg, South Africa, and is characterized by a compressed hollow thallus, pinnate with elongate, and linear-lanceolate branches. This species is distributed in South America, Africa, the Indian Ocean, Asia, Australia, and the Pacific Ocean (Guiry and Guiry 2013). Champia bifida was established by Okamura (1901) based on specimens collected from Enoshima, Japan, and is only distributed in the northwestern Pacific region (Guiry and Guiry 2013). Champia bifida is distinguished by having bifid apices, subdichotomous branches, and a broader size of segments than those of C. compressa (Okamura 1901).
Champia expansa was first described by Yendo (1903) based on specimens collected from Misaki, Japan. Despite a much different external appearance from the other members of the genus, he placed this species in Champia based on vegetative and reproductive structures. Yendo (1903) described that it was erect, compressed, dichotomously branched expanding in a flabellate manner, and had compressed segments due to much greater breadth than the length of segments. Okamura (1923) reported additional characteristics such as linear or cuneate segments of 6-10 mm breadth and a mucilaginous texture. This species has been reported only from Korea and Japan (Lee and Kang 2001, Guiry and Guiry 2013).
Champia parvula was originally described as Chondria parvula by C. Agardh (1824), and the type locality was Cadiz, Spain. This species has been reported from the most temperate and tropical coasts of the world (Reedman and Womersley 1976, Lozada-Troche and Ballantine 2010, Guiry and Guiry 2013). However, Irvine and Guiry (1983) speculated that this species is restricted to the eastern Atlantic and is not as widely distributed as reports indicate (Lozada-Troche and Ballantine 2010). This observation suggests that C. parvula reported from north-east Asia may represent a complex of species rather than a single entity (Irvine and Guiry 1983). In Korea, there are two morphological types of C. parvula confused when identifying species in the field; one has an entangled thallus and hooked apices, whereas the other has an erect thallus and straight apices.
Red algal systematists increasingly rely on molecular data to resolve species boundaries, as well as deep phylogeny (Le Gall and Saunders 2010, Yang et al. 2013). DNA barcoding, such as 5′ end of the cytochrome oxidase I mitochondrial gene (COI), in particular has proved useful for species-level identification of red algae (Saunders 2005, Kim et al. 2010). The COI-5P gene typically has none overlapping intraspecific vs. interspecific divergence. A character usually referred to as the “barcode gap” allows specimens to be assigned unambiguously as genetic species (Le Gall and Saunders 2010). Two new species, C. harveyana D.L. Ballantine & C. Lozada-Troche and C. puertoricensis Lozada-Troche & D.L. Ballantine, were discovered from the Caribbean Sea by molecular phylogenetic analyses and morphological characteristics (Ballantine and Lozada-Troche 2008, Lozada-Troche and Ballantine 2010). Champia harveyana and C. puertoricensis were separated from C. salicornioides Harvey and C. parvula, respectively.
In the present study, we tested the effectiveness of DNA barcoding for identifying and discovering Champia species diversity in Korea. We aimed to (i) confirm the delimitation of Champia species, (ii) reinstate the name C. bifida instead of C. compressa and (iii) describe a new species and new record, which previously identified as C. “parvula” in Korea, based on morphology and molecular analyses.
Specimens were collected from the subtidal and intertidal zones of Korea (Table 1). Samples used in morphological studies were preserved in 5% formalin/seawater and pressed on herbarium sheets. Voucher specimens were deposited in the herbarium of Jeju National University, Korea (JNUB). Thalli were sectioned using a hand or freezing microtome NK-101-II (Nippon Optical Works Co. Ltd., Tokyo, Japan). Sections were stained with 1% aniline blue acidified with 1% HCl and mounted on glass slides in 50% corn syrup solution. Photomicrographs were taken with a Q-imaging QICAM Fast (Burnaby, BC, Canada) digital camera mounted on an Olympus BX50 microscope (Olympus, Tokyo, Japan).
Genomic DNA was extracted from the silica-gel-dried specimens using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The gene selected to infer the phylogeny of the genus Champia was chloroplast-encoded rbcL and, to test the effectiveness of DNA barcoding, we used the mitochondrial-encoded COI gene. The primer pairs used for PCR amplification and sequencing reaction of the rbcL gene were rbcLF7-rbcLR753 and rbcLF645-rbcS start (Kim
and Kang 2011) and rbcLF145-rbcLR898 and rbcLF762-rbcLR1442 (Kim et al. 2012). For samples that failed amplification, we redesigned the primers (e.g., CrbcLF725 [5′-CGAAGACATGTATGAAAGAG-3′]andCrbcLR1407 [5′-CAGCAGTATCAGTAGAAGTA-3′]combination).The COI-5P region was amplified via PCR following Kim et al. (2010). Amplification condition for both rbcL and COI consisted of 5 min at 94˚C for denaturation, followed by 35 cycles of 1 min at 94˚C, 1 min at either 47˚C or 45˚C, and 2 min at 72˚C, with a final 7 min extension cycles at 72˚C, and a soak cycle at 4˚C. Resulting products were purified using the AccuPrep® PCR Purification Kit (Bioneer, Daejeon, Korea) following the manufacturer’s instructions.
We collated all sequences and aligned them visually using BioEdit ver. 7.1.11 (Hall 1999). Barcode data analyses were conducted in Mega ver. 5.10 (Tamura et al. 2011) with distance corrected under a Kimura 2-parameter model, and neighbor-joining was used to provide a visual display of COI-5P variation within and between species. Maximum likelihood (ML) analysis was conducted for phylogeny using RAxML software (Stamatakis 2006) and the GTR + Γ evolutionary model. We used 200 independent tree inferences using the -# option (200 distinct ML trees) with default –I (automatically optimized SPR rearrangement) and –c (25 distinct rate categories) options of the program to identify the best tree. We used the same program with the same setting and 1000 replications to generate bootstrap values for these phylogenies. Bayesian
analyses (BA) were conducted using MrBayes ver. 3.1.2 (Ronquist and Huelsenbeck 2003). Three million generations of two independent runs were performed with four chains for each matrix, and the burn-in period was identified graphically by tracking the likelihood at each generation to determine whether they had reached a plateau.
We obtained COI sequences of thirty-eight Champia specimens including C. parvula collected from the type locality in Spain, in addition to seven sequences from GenBank (Table 1). The amplified size of COI region ranged from 629 to 689 bp, depending on the primer combination used, of which we analyzed 616 bp corresponding to sequences from GenBank. An unrooted phylogram using neighbor-joining analysis showed the molecular distance within and between morphologically identified species (Fig. 1). Thirty-seven individuals of Korean Champia species resolved into four expected clusters that were assignable to Champia sp. (n = 7), C. recta (n = 10), C. bifida (n = 7), and C. expansa (n = 13). Four species showed interspecific divergences of 3.2-16.7%, whereas the respective mean intraspecific divergences were 0.1% in C. bifida, 0.7% in C. recta, 0.2% in Champia sp., and 0.02% in C. expansa. Champia sp. and C. recta were separated by 3.2-4.4% sequence divergence and also distinctly separated from C. parvula from Spain (CHA001) by 16.1-16.7% sequence divergence. Champia sp. and C. recta from Korea were allied with four Hawaiian C. parvula (HQ422763, HQ423089, HQ422822, HQ422864), but they showed topology independently with 4.4-7.6% sequence divergence (Sherwood et al. 2010). Champia bifida was separated by 12.8-14.3% sequence divergence from Hawaiian C. vieillardii (HQ422943, HQ422762, HQ423089), which has a compressed thallus (Sherwood et al. 2010). Champia expansa was clearly separated from other species by 12.2% (from C. bifida) to 15.5% (from Champia sp.) sequence divergences. Saunders and McDonald (2010) reported intraspecific divergence of three genera in the order Rhodymeniales: 0-2% in Rhodymenia, 0-0.9% in Halopeltis, and 0-0.75% in Pseudohalopeltis. In our study, the Korean Champia specimens showed 0-0.7% intraspecific divergence, which was within the range of the order Rhodymeniales (Saunders and McDonald 2010). The “barcoding gap” is defined as the difference between the maximum intraspecific and minimum interspecific divergence (Freshwater et al. 2010). The genus Champia from Korea had a barcoding gap of 1.7% maximum intraspecific and 3.2% minimum interspecific divergence in the COI marker. The results of our DNA barcoding analysis suggest that the mitochondrial-encoded COI gene is a useful marker for delimitating morphologically similar taxonomic groups in the genus Champia.
Champia inkyua Y. H. Koh, G. Y. Cho and M. S. Kim sp. nov.
Holotype: JNUB (CP120327-1), vegetative thallus, 27 March 2012 (Fig. 3A), deposited in the Herbarium of Jeju National University, Korea (JNUB).
Isotype: KB (NIBRAL0000137946)
Type locality: Jocheon, Jeju, Korea (33°32′41.57″N, 126°38′02.25″ E).
Etymology: The species epithet was chosen to honor the emeritus professor In Kyu Lee (Seoul National University) for his scientific achievements. He is the pioneer of marine macroalgal taxonomy in Korea. In particular, he made a great contribution to the order Rhodymeniales, Rhodophyta.
Korean name: 갈고리사슬풀
Description: Thallus entangled, hollow branches with terete segments constricted at septal regions, irregularly branched with hooked apices; B/L (Breadth/Length) ratio of segments between diaphragms is 1-1.5; cortex consists of irregularly shaped large and small cells; longitudinal filaments are running throughout the frond periphery and composed of a complete cell with two half cells between diaphragms in the cavity; gland cells are cut off from longitudinal filaments, spherical to ovoid in shape, 10-15 μm in diameter; tetrasporangia are forming from cortical cells, spherical in shape, tetrahedrally divided, 60-100 μm in diameter.
Morphology: Thalli are irregularly and radially bran- ched with several axes for 3 or 4 orders. Mature plant entangles itself to form a bush and grows up to 10 cm in diameter (Fig. 3A). Axes and branches are terete, 1-1.5 mm in diameter below, tapering gradually to branchlets of
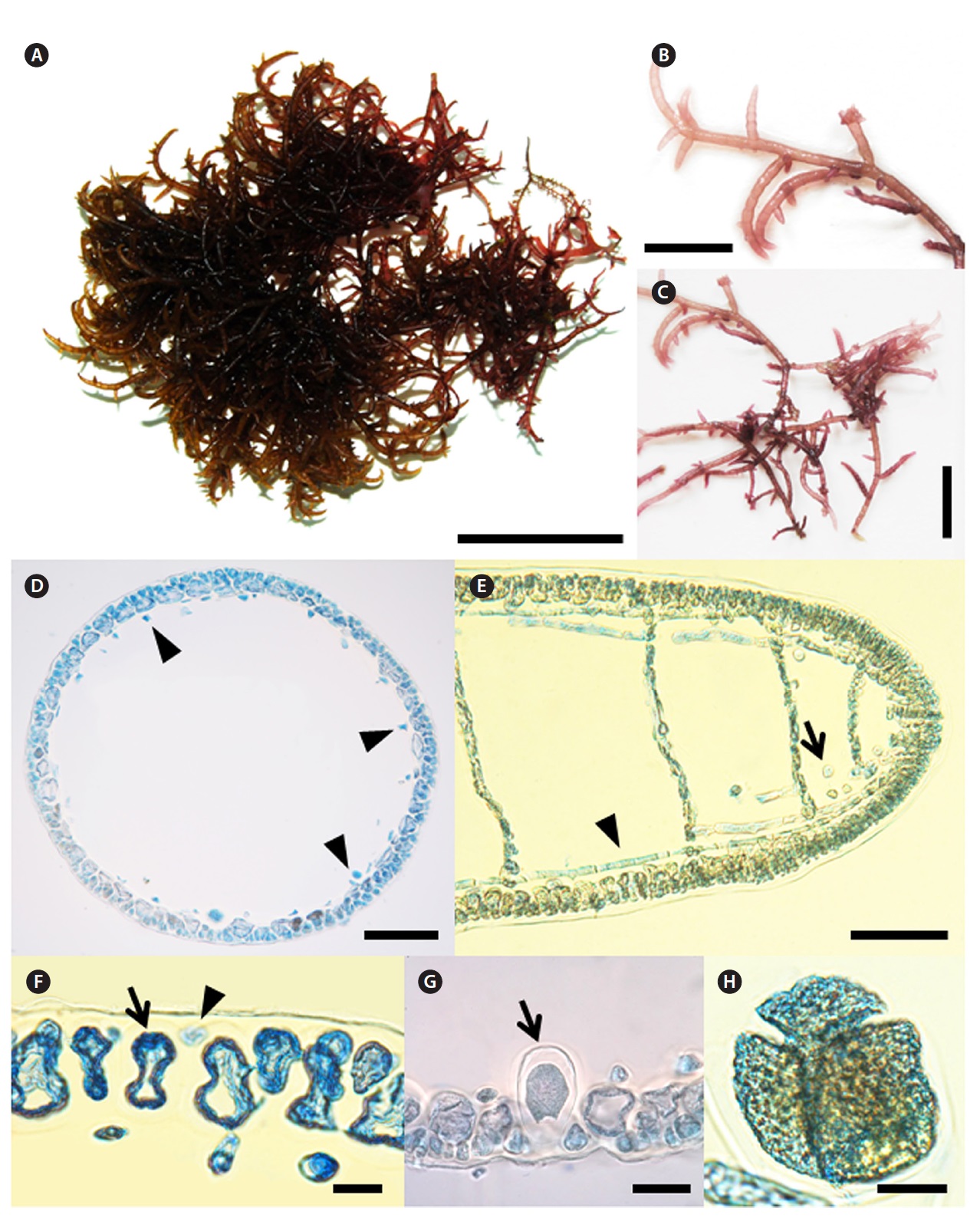
[Fig. 3.] Champia inkyua Y. H. Koh, G. Y. Cho and M. S. Kim sp. nov. (A) Holotype specimen (CP120327-1, vegetative, 27 March 2012, Jocheon, Jeju, Korea)
deposited at the Herbarium of Jeju National University (JNUB), Jeju, Korea. (B) Apical part of branch showing hooked apices. (C) Middle part of branch
showing irregular branching patterns. (D) Transverse section of thallus showing cut longitudinal filament cells (arrow head). (E) Longitudinal section of
thallus showing longitudinal filaments (arrow head) and gland cell (arrow). (F) Transverse section of thallus with large (arrow) and small cortical cell (arrow
head). (G) Tetrasporangia (arrow) placed cortical cells and enlarged toward inner hollow. (H) Mature tetrasporangia divided tetrahedrally. Scale bars: A, 3
cm; B, C, 1 cm; D, E, 100 μm; F, G, H, 20 μm.
0.3-0.5 mm diameter. Branches are slightly constricted at diaphragms and ends are hooked or straight (Fig. 3B and 3C). After a transverse division of each apical cell, the lower cell divides longitudinally to make an inner cell and an outer cell, from which filaments and cortical cells are produced, respectively (Fig. 3E). Segments in the middle part of the axis are up to 0.3 mm broad and 0.2 mm long. The B/L ratio of the segments was about 1-1.5 (Fig. 3B and 3C). Cortex consists of one-layer of large cells and randomly occurring small cells cut off obliquely from the large cells. The large cells are 30-45 μm long, whereas the small cells are 7-15 μm long (Fig. 3F). Longitudinal filaments of the medulla run through the inner hollow. Filaments elongated longitudinally and laterally (Fig. 3D and 3E). Filaments at the upper part of the thallus are 8-12 μm broad and 50-80 μm long. Globular gland cells are produced singly on filaments and are 10-15 μm in diameter (Fig. 3E). Diaphragms are distinct throughout most of the thallus (Fig. 3B, 3C, and 3E) but are obscured near older bases. Tetrasporangia are scattered and transformed from cortical cells, 60-100 μm in diameter, tetrahedrally divided (Fig. 3H). Plants are epiphytic on other macroalgae forming an entangled bush and grow in the tidal pool of the intertidal zone.
Remarks: This species has been identified as C. parvula from Korea (Lee and Kang 2001). Champia parvula was characterized with an erect and spreading thallus, terete branches irregularly with straight apices, longitudinal filaments composed of two (rarely three) complete cells and two part cells between the diaphragms (Harvey 1853, Womersley 1996). We examined the morphology of C. parvula collected from San Pedro de Veigue, Spain, near the type locality as a comparison with Korean specimens (Harvey 1853, Reedman and Womersley 1976, Womersley 1996). As a result, C. inkyua sp. nov. from Korea showed morphological similarities with C. parvula from Spain, but C. inkyua has an entangled thallus, hooked apices, and is composed of a complete cell with two half cells between diaphragms in the cavity, and large cortical cells (Reedman and Womersley 1976, Womersley 1996). Lee (1978) described C. parvula collected from Hokkaido, Japan as having terete axes and irregular branches, but C. parvula was absent of hooked apices and developed longitudinal filaments in peripheral with central. As a terete species with irregular branches, C. affinis (Hooker and Harvey) Harvey from Australia is also close to C. inkyua, but can be distinguished by outer cortical development in C. affinis (Reedman and Womersley 1976). Champia recta Noda has similar morphological features with C. inkyua (Noda 1973). However, C. recta can be distinguished by its erect and pyramidal growth form with an absence of hooked branches (Noda 1973). In the DNA barcoding result, C. inkyua was clearly separated from Spanish C. parvula collected from their type locality by 16.1-16.5% and Hawaiian C. parvula by 5.1-7.6% sequence divergence (Sherwood et al. 2010). In addition, C. inkyua was distinguished from C. recta, which formed a sister clade, by 3.2-4.4% sequence divergence. In the rbcL analysis, C. inkyua was distinctly separated from C. parvula from Spain by 10.9-11.1% sequences divergence. Other Champia species having terete branches were clearly separated from C. puertoricensis by 10.1-10.3%, C. salicornioides by 12-13.2%, and C. harveyana by 11.4-11.9% sequences divergence (Lozada-Troche and Ballantine 2010, Suzuki et al. 2010). Champia inkyua and C. recta formed a sister clade, but they were separated by 1.7-2% divergence. Although the interspecific divergence was lower than that of the others, they had different morphological characteristics. Therefore, we suggest a new species, C. inkyua, with hooked branch apices and entangled branches to form bush entity, instead of C. parvula identified previously in Korea.
Champia recta Noda 1973
Type locality: Banjin, Kashiwazaki, Niigata Prefecture, Japan.
Korean name: 참사슬풀
Morphology: Thalli are erect, attached by a disc-like holdfast, 3-5 cm high, main axes teret, about 1 mm thick, and profusely branched throughout. Branches are single or verticilate, decreasing in length in a pyramidal appearance, not attenuate at the base. Apices are slightly attenuated, beset with secondary branchlets mostly single (Fig. 4B and 4C). Fronds are septated throughout. Internodes are more or less barrel-shaped. The segments are subequral to or about 1.5 times as long as the diameter (Fig. 4C-4E). Segments are up to 0.3 mm broad and 0.2 mm long at the middle part of the axis. B/L ratio of the segments is about 1-1.5 (Fig. 4D). Internal frond is composed of a cortex of small cells, large cells within. The large cells are 25-45 μm long, whereas the small cells are 7-12 μm long. Longitudinal filaments of the medulla are transverse through the inner hollow. The filaments are elongate longitudinally and laterally. Filaments at upper part of thallus are 5-7 μm broad and 50-110 μm long (Fig. 4G). Diaphragms are developed by one-layer of cells that regularly transverse the inner hollow to make the plant segmented (Fig. 4G). Tetrasporangia are formed just below the surface of the upper branches; spherical to ovoid. They are tetrahedrally divided and 50-60 μm broad and 65-75 μm
long (Fig. 4H). Plants grow on rocks or are epiphytic on other macroalgae in the lower intertidal zone.
Remarks: Champia recta was first reported by Noda (1973) based on specimens collected from Banjin-misaki, Japan. Champia recta was characterized by the main axes terete, single or verticillate branches, and decreasing in length in a pyramidal appearance (Noda 1973). However, Yoshida (1998) mentioned that the diagnostic characters of C. recta were within the variation range of C. parvula, and synonymized the name C. recta. Results of the morphological comparison between C. recta from Korea and C. parvula from Spain showed a difference in the size of the thallus, cortical cells, and longitudinal filament composition (Reedman and Womersley 1976, Womersley 1996). In the DNA barcoding result, C. recta was clearly separated from Spanish C. parvula by 16.3-16.7% and from Hawaiian C. parvula by 4.4-5.7% sequence divergence (Sherwood et al. 2010). Champia recta was clearly
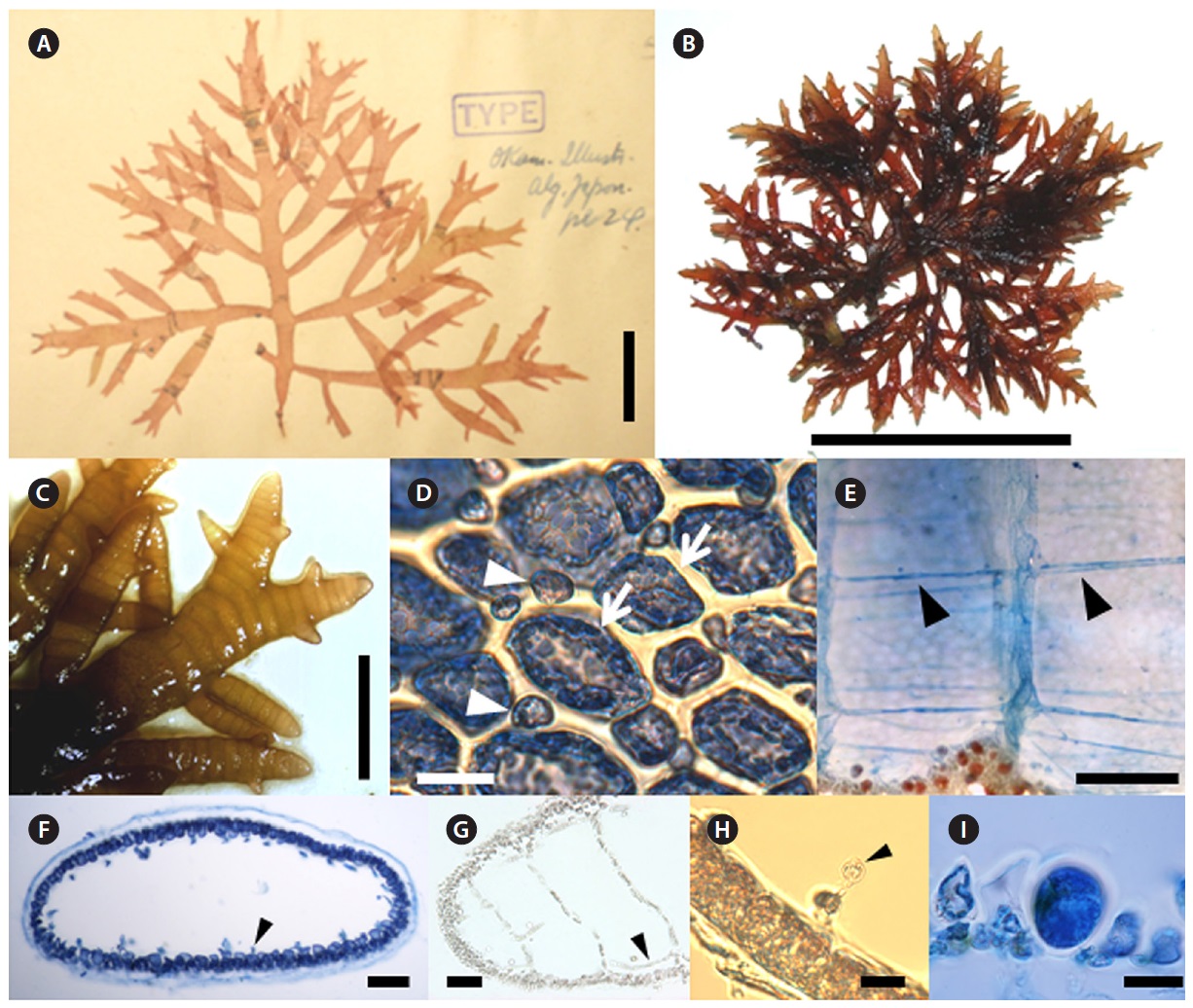
[Fig. 5.] Champia bifida Okamura. (A) Photo of holotype specimen deposited at the Herbarium of Hokkaido University (SAP), Japan (photo taken by M.S.
Kim). (B) Tetrasporophyte collected from Bukchon on 22 March 2012. (C) Apical part of branches showing bifid apex. (D) Surface view of cortex with large
(arrow) and small cells (arrow head). (E) Longitudinal filaments are running throughout the center of inner hollow (arrow head). (F) Transverse section of
thallus showing cut longitudinal filament cells (arrow head). (G) Longitudinal section of thallus with longitudinal filament (arrow head). (H) Transverse
section of thallus with glandular cell (arrow head) connected with cut longitudinal filament. (I) Tetrasporangia placed cortical cells and enlarged toward
inner hollow. Scale bars: A, 2 cm; B, 5 cm; C, 3 cm; D, H, 20 μm; E, 500 μm; F, 100 μm; G, I, 50 μm.
distinguished from C. inkyua by 3.2-4.4%. In the rbcL analysis, C. recta was distinctly separated from C. parvula from Spain by 11.5-11.6% sequences divergence. Other Champia species having terete branches were clearly separated from C. puertoricensis by 10.7-10.8%, C. salicornioides by 12.7-13.6%, and C. harveyana by 12.1-12.3% sequences divergence (Lozada-Troche and Ballantine 2010, Suzuki et al. 2010). In addition, the Korean entity in the rbcL analysis was almost identical with Japanese specimens previously identified as C. parvula (personal communication, data not shown). Therefore, we reinstate the name C. recta for the entity of C. parvula identified previously in Korea based on morphological and molecular evidence of an erect thallus and straight apices.
Champia bifida Okamura 1901
Type locality: Enoshima and Misaki, Kanagawa Prefecture, Japan.
Korean name: 두갈래사슬풀
Morphology: Thalli are rather prostrate or erect and always very iridescent when living, tubularly-compressed and consist of a prominent axis and lateral branches. The branches are pinnate or alternate to expand on the same plane. Second or third orders of branches also show a similar pattern. Fully grown thallus is up to 5 cm high, their axes up to 2.5 mm broad, and some branches have bifid apices (Fig. 5B and 5C). After a transverse division of each apical cell, the lower cell divides longitudinally to make an inner cell and an outer cell, from which filaments and cortical cells are produced, respectively (Fig. 5G). The cortex consists of one-layer of large cells and randomly occurring small cells cut off obliquely from the large cells. Large cells are 20-45 μm long, whereas the small cells are 7-20 μm long (Fig. 5D). Longitudinal filaments of the medulla run through the inner hollow (Fig. 5E-5G). Filaments at the upper part of the thallus are 5-9 μm broad and 70-95 μm long. Globular gland cells are produced singly on filaments and are 5-10 μm in diameter (Fig. 5H). Diaphragms are developed by one-layer of cells and regularly transverse the inner hollow to make the plant segmented (Fig. 5E and 5G). Tetrasporangia are among the cortical cells and are enlarged toward the inner hollow. They are 30-40 μm broad and 50-60 μm long (Fig. 5I). Plants grow in the lower intertidal to subtidal (1-5 m) zone on rock or other algae.
Remarks: Champia bifida and C. compressa were confused in Korea by similar morphological features such as shape of thallus, branching patterns, and growth form (Park and Lee 1998). Okamura (1901) described the bifid apex, the subdichotomous branches, and the broader size of segments as the diagnostic characters of C. bifida. Park and Lee (1998) concluded that the Japanese and Korean plants known as C. bifida should be identified as C. compressa, based comparing two type specimens. Champia compressa was characterized by a compressed thallus, pinnate with elongate and lineari-lanceolate branches (Agardh 1876). Subsequent studies pointed out that the branching pattern, B/L ratio, and compressed thallus were diagnostic characters of the species (Millar 1990, Masuda et al. 2001, De Clerck et al. 2005). Millar (1990) remarked that the compressed thalli with pinnate branching pattern of C. compressa are superficially similar to the New Caledonian species, C. vieillardii Kützing. However, C. vieillardii has much flattened branches, and longitudinal filaments are found mostly near the periphery of the diaphragms rather than in the centres (Millar 1990). Korean specimens had a bifid apex, and segment sizes were broader than those of C. compressa (Millar 1990, Masuda et al. 2001, De Clerck et al. 2005). However, Park and Lee (1998) reported that the diagnostic characters of C. bifida were within the variation range of C. compressa. In the DNA barcoding result, C. bifida was clearly separated from other Champia species by 12.2-16.7% and from Hawaiian C. vieillardii with a compressed thallus by 11.9-14.1% sequence divergence (Sherwood et al. 2010). In the rbcL analysis, Korean specimens were clearly separated from Floridian (AY294358) and Australian (HQ400605) C. compressa by 9.2% from each other, and from Puerto Rican C. vieillardii by 6.9-7.3% (Lozada-Troche and Ballantine 2010). In addition, Korean entities were almost identical with Japanese C. bifida collected from their type locality (personal communication, data not shown). Thus, we reinstate the name C. bifida instead of C. compressa previously identified in Korea based on morphological and molecular evidence.
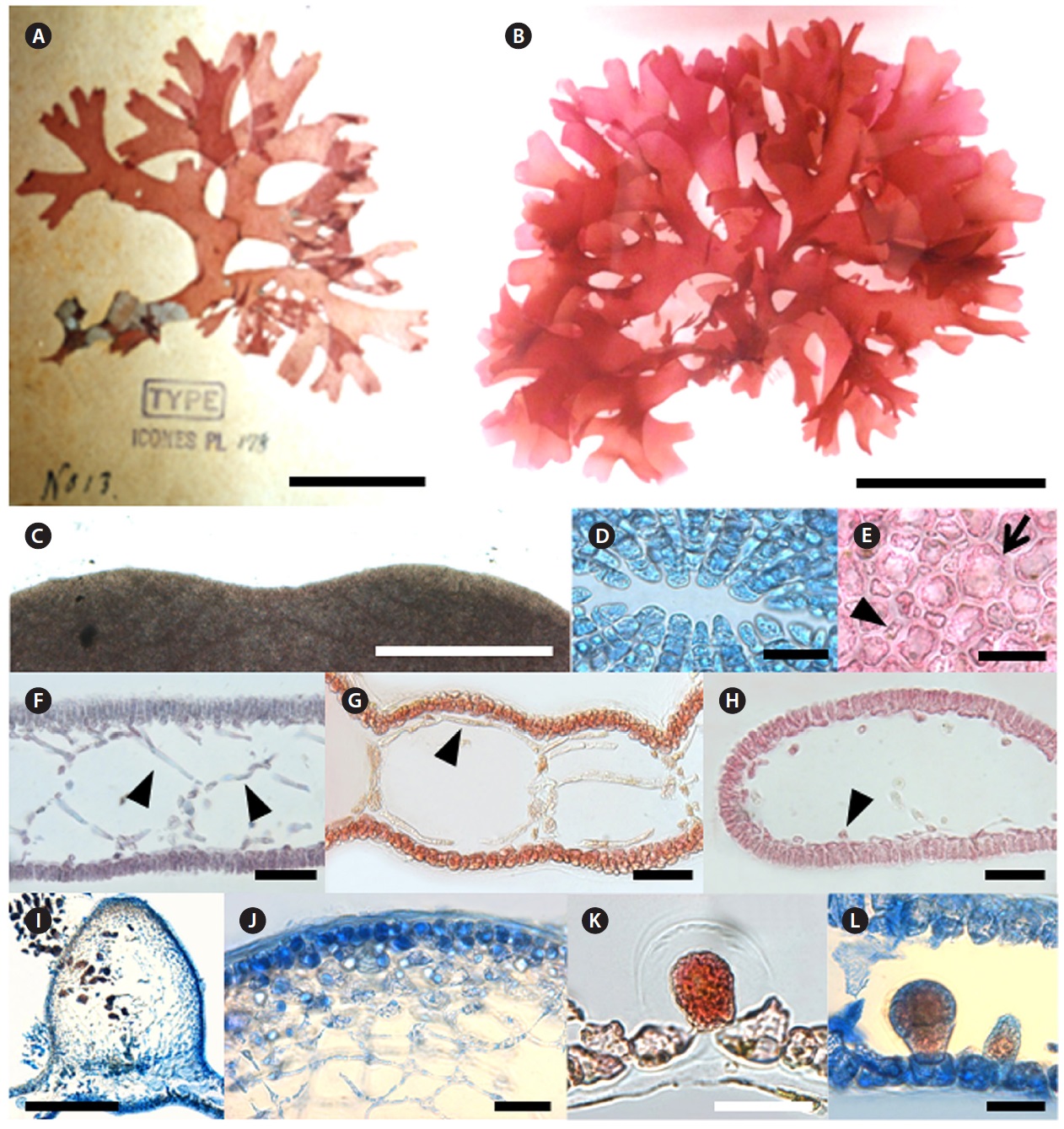
Champia expansa Yendo 1903
Type locality: Misaki, Kanagawa Prefecture, Japan.
Korean name: 넓은사슬풀
Morphology: Thalli are erect, branched dichotomously or subdichotomously, up to 3.5-9 cm in length, expanding into a compressed flabellum and mucilaginous (Fig. 6B). Apices are retuse (Fig. 6C) with several apical cells 5-9 μm length (Fig. 6D). Segments become 11 mm broad and 1.1 mm long at the middle part of plant. The B/L ratio of each segment is 7.8-9.1 (Fig. 6B and 6G). Apical cells divide transversely, and then the resulting lower cell undergoes an uneven longitudinal division to produce large inner cells and small outer cells. The inner cell becomes a filament or a diaphragm cell of the medulla, whereas the outer cell grows to a large cortical cell of 19-43 μm length which frequently cuts off small cells of 8-11 μm in length (Fig. 6E). Several filamentous cells form a net-like structure on the upper part of the plant (Fig. 6F). Longitudinal filaments of the medulla run through the inner hollow and become 5-10 μm broad and 70-110 μm long (Fig. 6G). Filaments are lateral, and branches are connected by a pit (Fig. 6H). Diaphragms are formed regularly in the inner hollow to make the plant segmented in outer appearance (Fig. 6G). Cystocarps are urceolate with a conspicuous ostiole and elevated from the surface at 1-1.2 mm height (Fig. 6I). Inner cells of the mature pericarp are stellate (Fig. 6J). Tetrasporangia are scattered mostly on one side of the surface. Tetrasporangium originated from a large cell of the cortex, grows toward the inner part of the thallus, and is divided tetrahedrally (Fig. 6K and 6L). Tetrasporangia are 50 μm broad, 75 μm long (Fig. 6L). Plants grow on rocks in the 5-10 m subtidal depth range.
Remarks: Korean C. expansa was clearly separated from other species of Champia based on their specific morphological features such as flabellate thallus, branching patterns, and density of marginal proliferations (Yendo 1903). Millar (1990) described the C. expansa collected from Coffs Harbour, New South Wales, Australia. He remarked that the specimens from Coffs Harbour and Japan differed only in overall size and cortical construction. The Australian samples had one layer of similar sized cortical cells, whereas the Japanese plants had two layers composed of two different sized cells (Millar 1990). Millar (1998) separated Australian C. womersleyi from Japanese C. expansa based on morphological characters such as the rare occurrence of proliferous in the blade margin and hairs that mimic carpogonia with trichogynes. Korean C. expansa had two layers composed of two different size cells, common occurrence of proliferous, mature thallus length up to 10 cm and lack of hairs. In the DNA barcoding result, C. expansa from Korea formed a clade almost identically and clearly separated from other Champia species by 11.8-15.7% sequence divergence. In the rbcL analysis,
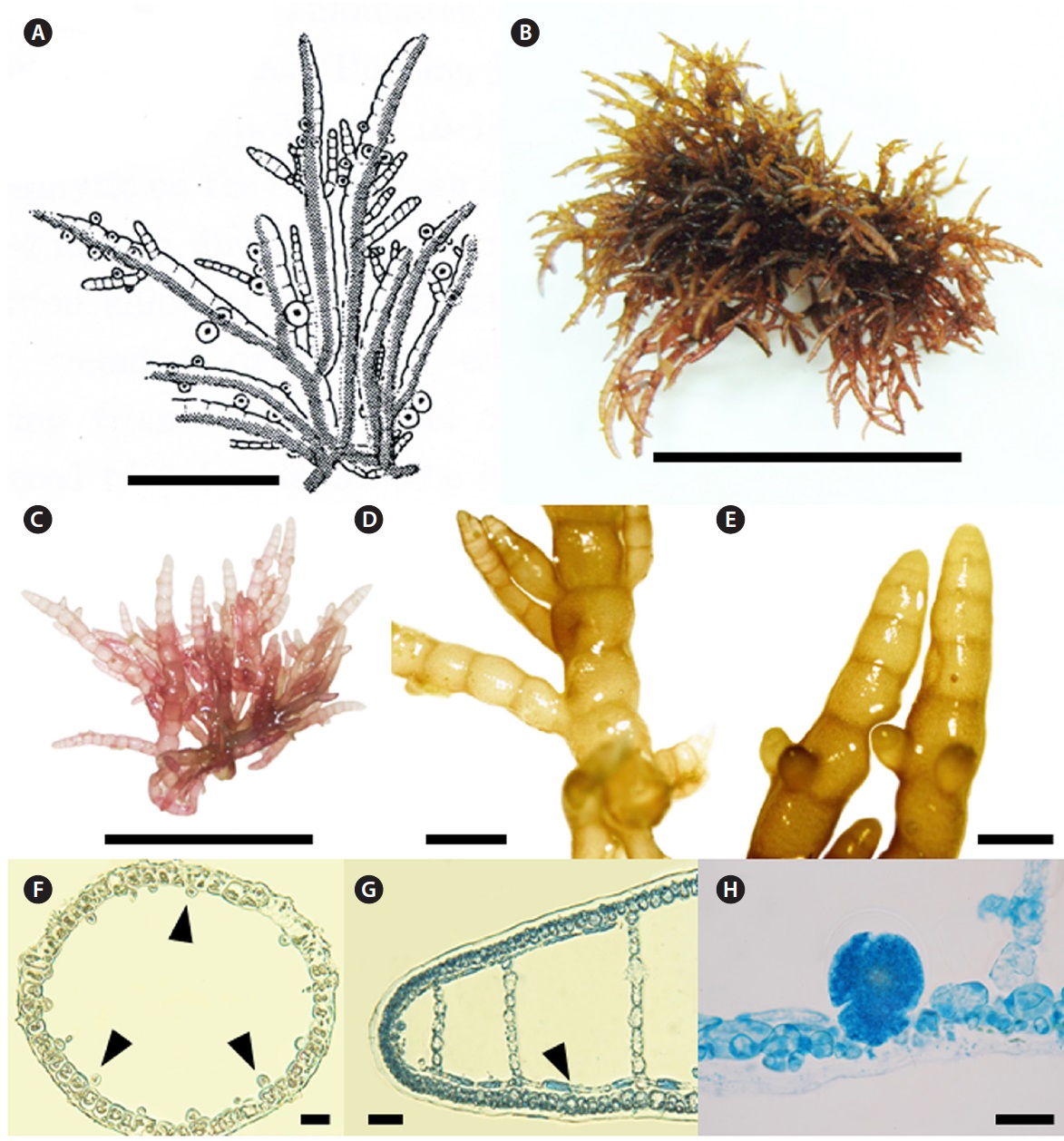
[Fig. 6.] Champia expansa Yendo. (A) Photo of holotype specimen deposited at the Herbarium of Hokkaido University (SAP), Japan (photo taken by
M.S. Kim). (B) Vegetative plant collected from Pyoseon on 11 March 2011. (C) Apical part of thallus showing retuse apex. (D) Apical view showing ring
grouped cells. (E) Surface view of cortex with large (arrow) and small cells (arrow head). (F) Transverse section of apical part showing several filamentous
cells forming net-like structure (arrow head). (G) Longitudinal section of thallus with longitudinal filaments (arrow head). (H) Transverse section of thallus
showing cut longitudinal filament cells (arrow head). (I) Longitudinal section of cystocarp. (J) Pericarp with stellate cells. (K-L) Tetrasporangia placed cortical
cells and enlarged toward inner hollow. Scale bars: A, 4 cm; B, 5 cm; C, I, 500 μm; D, E, J, K, L 50 μm; F, G, H, 100 μm.
C. expansa was separated from Australian C. compressa by 6.6-6.7% and from other species of Champia by 6.4-13.3% sequence divergence (Sherwood et al. 2010). In addition, Korean entities were almost identical with Japanese C. expansa collected from their type locality (personal communication, data not shown). DNA barcoding and rbcL analyses of C. expansa strongly supported their unique morphological characters.